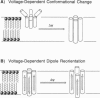
Abstract
A nitroxide spin label attached to the C-terminus of the channel forming peptide alamethicin produces an enhancement of the nuclear spin-lattice relaxation rates of peptide protons as a result of both intermolecular and intramolecular magnetic dipole-dipole interactions. The intermolecular contribution provides evidence that alamethicin monomers collide preferentially in a C-terminal-to-N-terminal configuration in methanol. From the intramolecular paramagnetic enhancement of nuclear spin-lattice relaxation times, effective distances between the unpaired electron on the nitroxide at the C-terminus of alamethicin and protons along the peptide backbone were calculated. These distances are much shorter than distances based on the reported crystal structure of alamethicin, and cannot be accounted for by motion in the bonds that attach the nitroxide to the peptide. In addition, the differences between distances deduced from the nuclear spin relaxation and the distances seen in the crystal structure increase toward the N-terminal end of the peptide. The simplest explanation for these data is that the alamethicin backbone suffers large structural fluctuations that yield shorter effective distances between the C-terminus and positions along the backbone. This finding can be interpreted in terms of a molecular mechanism for the voltage-gating of the alamethicin channel. When the distances between a paramagnetic center and a nucleus fluctuate, paramagnetic enhancements are expected to yield distances that are weighted by r-6, and distances calculated using the Solomon-Bloembergen equations may more nearly represent a distance of closest approach than a time average distance. Therefore, the use of paramagnetic centers such as spin labels or metal ions with long electron T1 values provides a distance measurement that reflects a dynamically averaged structure where the averaging process heavily weights short distances. The results of such measurements, when combined with other structural information, may provide particularly clear evidence for the magnitude of structural fluctuations involving distances greater than 10 A.
Full text
PDF
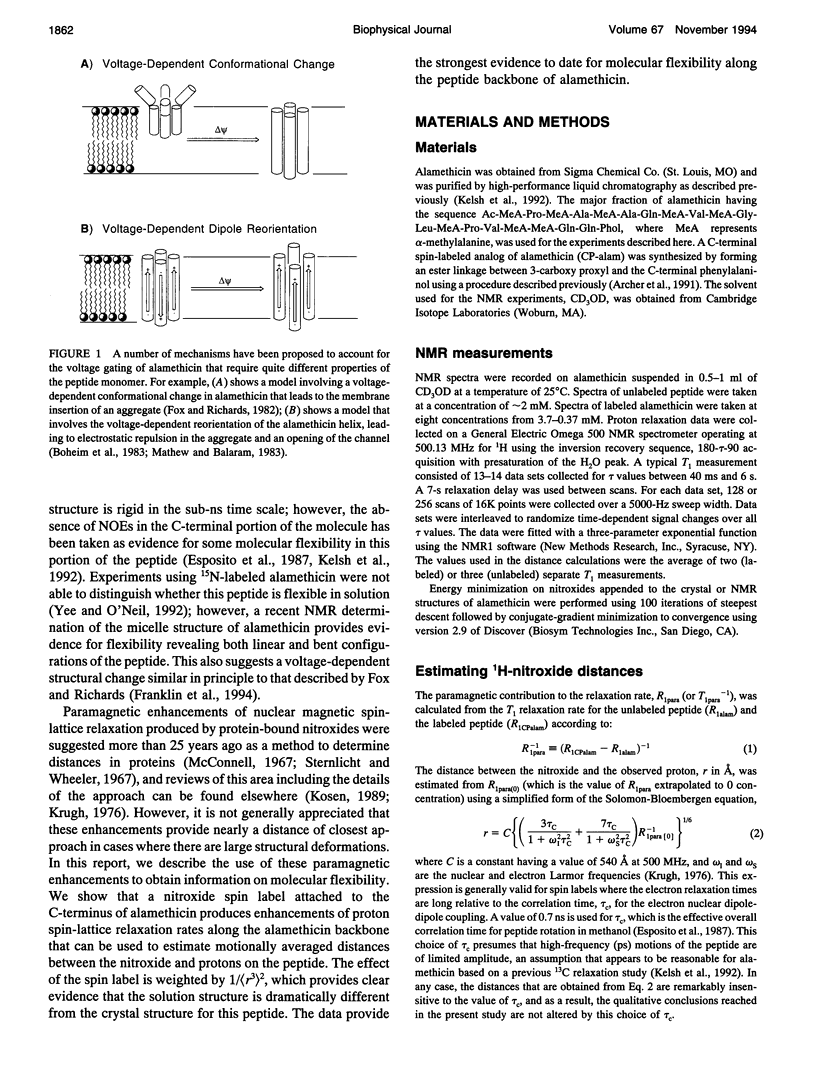
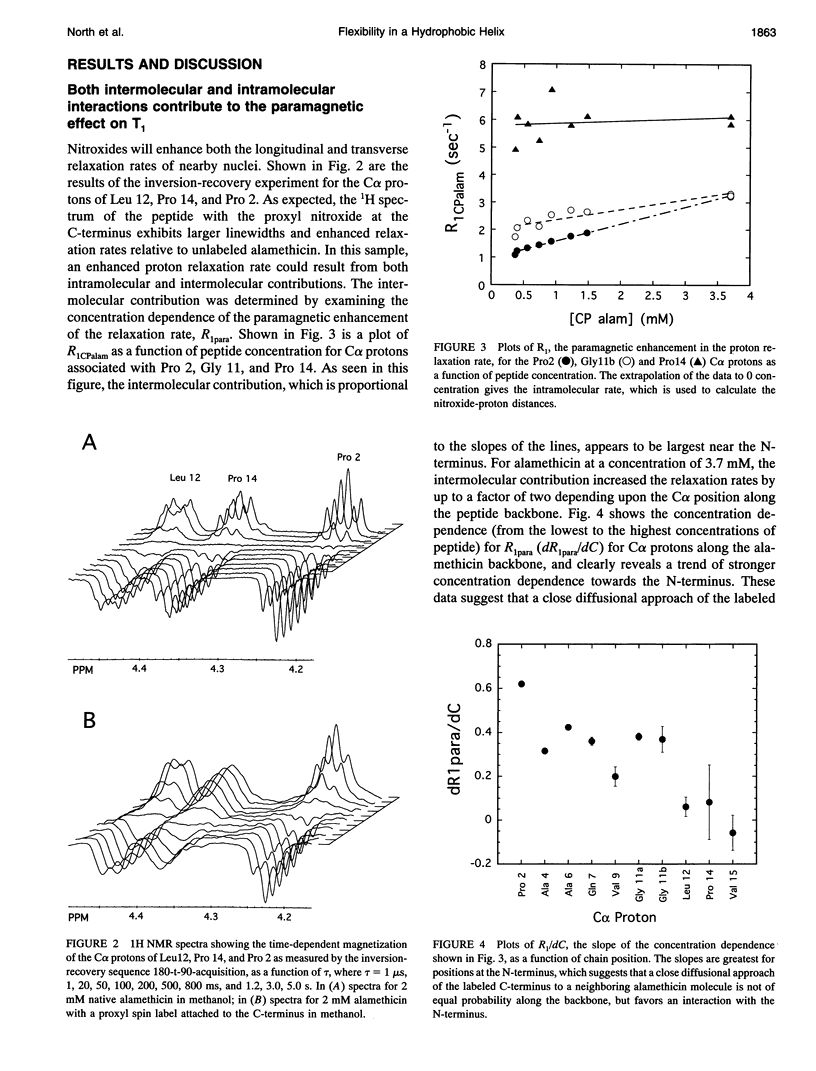
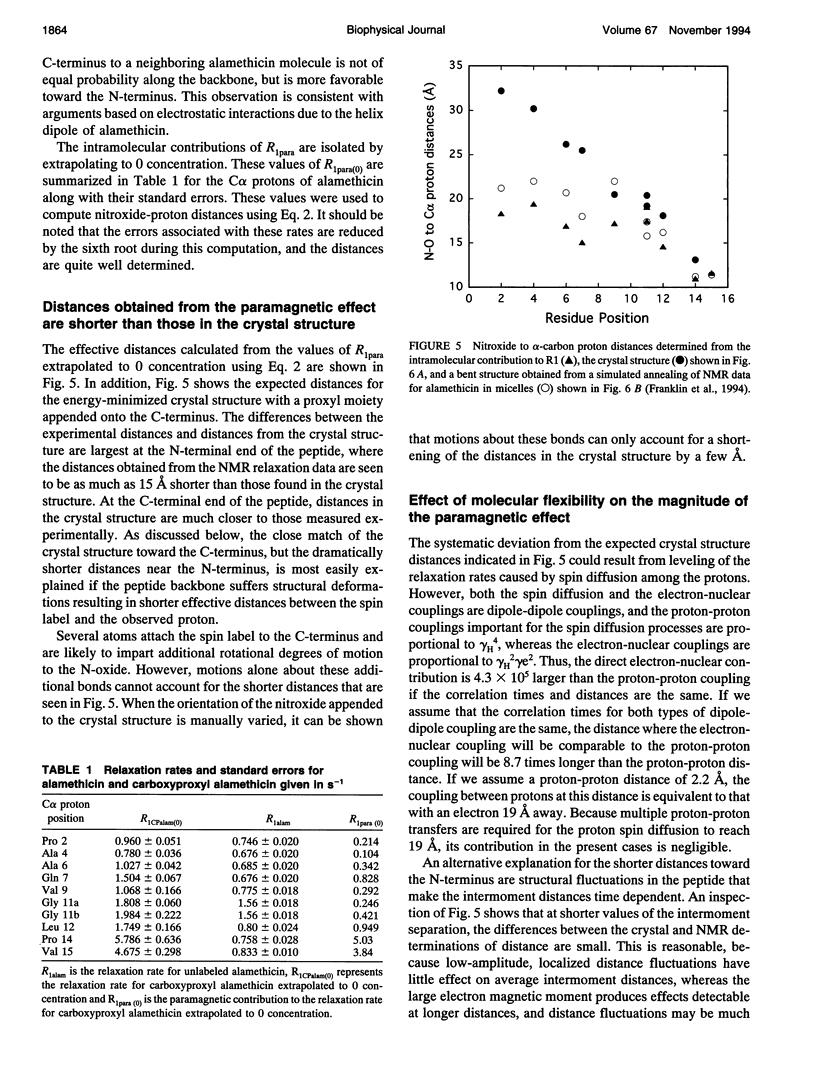


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer S. J., Ellena J. F., Cafiso D. S. Dynamics and aggregation of the peptide ion channel alamethicin. Measurements using spin-labeled peptides. Biophys J. 1991 Aug;60(2):389–398. doi: 10.1016/S0006-3495(91)82064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992 Jun 16;31(23):5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- Cafiso D. S. Alamethicin: a peptide model for voltage gating and protein-membrane interactions. Annu Rev Biophys Biomol Struct. 1994;23:141–165. doi: 10.1146/annurev.bb.23.060194.001041. [DOI] [PubMed] [Google Scholar]
- Esposito G., Carver J. A., Boyd J., Campbell I. D. High-resolution 1H NMR study of the solution structure of alamethicin. Biochemistry. 1987 Feb 24;26(4):1043–1050. doi: 10.1021/bi00378a010. [DOI] [PubMed] [Google Scholar]
- Fox R. O., Jr, Richards F. M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-A resolution. Nature. 1982 Nov 25;300(5890):325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- Franklin J. C., Ellena J. F., Jayasinghe S., Kelsh L. P., Cafiso D. S. Structure of micelle-associated alamethicin from 1H NMR. Evidence for conformational heterogeneity in a voltage-gated peptide. Biochemistry. 1994 Apr 5;33(13):4036–4045. doi: 10.1021/bi00179a032. [DOI] [PubMed] [Google Scholar]
- Fraternali F. Restrained and unrestrained molecular dynamics simulations in the NVT ensemble of alamethicin. Biopolymers. 1990;30(11-12):1083–1099. doi: 10.1002/bip.360301109. [DOI] [PubMed] [Google Scholar]
- Kelsh L. P., Ellena J. F., Cafiso D. S. Determination of the molecular dynamics of alamethicin using 13C NMR: implications for the mechanism of gating of a voltage-dependent channel. Biochemistry. 1992 Jun 9;31(22):5136–5144. doi: 10.1021/bi00137a007. [DOI] [PubMed] [Google Scholar]
- Mathew M. K., Balaram P. Alamethicin and related membrane channel forming polypeptides. Mol Cell Biochem. 1983;50(1):47–64. doi: 10.1007/BF00225279. [DOI] [PubMed] [Google Scholar]
- Post C. B. Internal motional averaging and three-dimensional structure determination by nuclear magnetic resonance. J Mol Biol. 1992 Apr 20;224(4):1087–1101. doi: 10.1016/0022-2836(92)90471-u. [DOI] [PubMed] [Google Scholar]
- Yee A. A., O'Neil J. D. Uniform 15N labeling of a fungal peptide: the structure and dynamics of an alamethicin by 15N and 1H NMR spectroscopy. Biochemistry. 1992 Mar 31;31(12):3135–3143. doi: 10.1021/bi00127a014. [DOI] [PubMed] [Google Scholar]