Abstract
Little is known about the nature of genetic variation underlying complex diseases in humans. One popular view proposes that mapping efforts should focus on identification of susceptibility mutations that are relatively old and at high frequency. It is generally assumed—at least for modeling purposes—that selection against complex disease mutations is so weak that it can be ignored. In this article, I propose an explicit model for the evolution of complex disease loci, incorporating mutation, random genetic drift, and the possibility of purifying selection against susceptibility mutations. I show that, for the most plausible range of mutation rates, neutral susceptibility alleles are unlikely to be at intermediate frequencies and contribute little to the overall genetic variance for the disease. Instead, it seems likely that the bulk of genetic variance underlying diseases is due to loci where susceptibility mutations are mildly deleterious and where there is a high overall mutation rate to the susceptible class. At such loci, the total frequency of susceptibility mutations may be quite high, but there is likely to be extensive allelic heterogeneity at many of these loci. I discuss some practical implications of these results for gene mapping efforts.
Introduction
Mapping the genes that contribute to complex diseases—such as diabetes, schizophrenia, and hypertension—will be a major challenge of the postgenome era (Risch 2000). Currently, little is known about the nature of genetic variation underlying complex diseases in humans, which makes it difficult to be confident about strategies for this problem. One popular hypothesis proposes that the genetic factors underlying common diseases will be alleles that are themselves quite common in the population at large (Lander 1996; Chakravarti 1999).
Assumptions about the genetic factors contributing to complex diseases are important in several ways (Zwick et al. 2000). Under the “common disease, common variant” hypothesis, it may be possible to create a catalogue of common SNPs and to use association mapping to identify disease-susceptibility mutations from that list (Risch and Merikangas 1996; Cargill et al. 1999; Halushka et al. 1999). Susceptibility mutations might also be detected indirectly through linkage disequilibrium with genotyped markers (Kruglyak 1999). A critical assumption of both association and linkage-disequilibrium mapping is that there is little allelic heterogeneity within loci. If a gene contains low-frequency mutations at many different sites—as is often the case in Mendelian disorders (Terwilliger and Weiss 1998; Green et al. 1999)—then the power of current statistical tests of association will be greatly reduced (Slager et al. 2000).
Assumptions about the likely frequency spectrum of disease mutations are also critical in modeling linkage disequilibrium (Kruglyak 1999; Long and Langley 1999; Zöllner and von Haeseler 2000). In particular, there tends to be more linkage disequilibrium around rare mutations (Kruglyak 1999). This has implications both for the density of markers that will be needed to scan a region for associations and for the problem of designing optimal statistical tests.
In this article, I propose a model for the evolution of genetic variation underlying a complex disease. This model is used to predict various properties of susceptibility mutations that will be important for designing mapping strategies. The model contains various simplifications, and the parameter values are not known accurately, so the conclusions of the article should be viewed as qualitatitive. Nonetheless, it seems that theoretical models can be a useful guide at this early stage of the search for complex-disease genes.
The Results section of this article describes three aspects of the proposed model. I begin by focusing on the overall frequency of susceptibility alleles (1) at a single complex-disease locus and (2) assuming a specific (multiplicative) model of interactions among loci. I then examine the properties of the independent mutations that combine to make up the class of susceptibility alleles at a locus.
Models and Assumptions
Unlike Mendelian traits, which are controlled by genes of large effect and show simple patterns of inheritance within families, the transmission of complex phenotypes is governed by multiple factors, and familial patterns of inheritance are complicated. Phenotypic outcomes may be determined by a mixture of genetic factors (i.e., variation at multiple loci) plus environmental and stochastic factors. A defining feature of complex phenotypes is that no single locus contains alleles that are necessary or sufficient for disease. (For some complex traits, there are also rare Mendelian forms, but these can be considered separately from the more common, “complex” forms of the diseases.)
Before genetic studies of a complex trait are conducted, it is common to estimate the increase in risk among relatives of an individual affected with the disease of interest; this gives some indication of the overall magnitude of genetic effects. Assuming a binary phenotype, let K be the population frequency of the disease, and let Kr be the probability that an r-degree relative of an affected proband is also affected. Here, I will focus on the sibling recurrence ratio Λs, defined as Ks/K, where Ks is the sibling recurrence risk. Ideally, Λs might be estimated using siblings reared apart, so that any increased risk is due to genetic factors alone with no contribution from environmental correlations.
A pair of articles by Risch (1990a, 1990b) describe multilocus models of inheritance for a complex disease and show how to calculate Λs on the basis of allele frequencies and penetrance values at each locus. It is shown that Λs can be partitioned into single-locus components (and possibly higher-order terms), and that the magnitudes of the single-locus components are critical in determining the power of affected sib-pair studies.
The models of Risch (1990a) provide a convenient starting point for the present study. Specifically, I consider a binary trait and assume that the probability that an individual is affected (i.e., the penetrance) depends on his or her genotype at L genes. In this model, the role of nongenetic factors is not considered explicitly but enters the model implicitly, in the sense that the penetrance of a given genotype can be thought of as a weighted average over the possible environmental states. As shown by Risch (1990a), it is convenient to assume that penetrance factors are multiplicative across loci. This corresponds to a class of models with epistasis. In one subsection of the Results, “Multilocus Models,” it will be necessary to make specific assumptions about the penetrance model. There are few relevant biological data to guide us here, so the choice to use a multiplicative model is based largely on simplicity and convenience. It is hoped that the results can nonetheless be useful in guiding our intuition. The implications of these particular assumptions are discussed in more detail below.
In Risch (1990a), as is customary in theoretical work relating to complex diseases, the allele frequencies at each of the disease loci are treated as parameters of the model. The approach that I take here is very different—namely, the allele frequencies are treated as random, resulting from an evolutionary process including selection, mutation, and genetic drift. The goal of modeling the evolutionary process is to learn about the underlying allele-frequency distributions.
During construction of the evolutionary model, it is important to think carefully about the role of natural selection. Of course, some mutations that increase disease susceptibility—particularly for diseases that occur late in life, after reproduction—may experience little or no selection against them. The model outlined below will allow for this possibility. However, it is plausible that even mutations whose primary effect occurs late in life may also have a weak deleterious effect early in life. For example, a mutation that predisposes individuals to Alzheimer disease might also cause subtle changes in brain function early in life. I will show that even very small selection coefficients, of the order of 10-4, can affect the frequency distribution of an allele, even though an effect of this size would be virtually impossible to measure directly. These considerations suggest that there may be no simple relationship between the selection coefficients and the penetrance values for a given mutation. Here, I model the selection and penetrance values as being independent.
In this study, I focus on a model of purifying selection. It should be noted however, that some loci may be subject to balancing selection, which has rather different consequences for allele frequencies. However, evolutionary studies of patterns of genetic variation in humans and other organisms suggest that balancing selection is relatively infrequent compared with purifying selection (e.g., Przeworski et al. 2000), except in the MHC region. I will not consider balancing selection further here.
Specific Assumptions
First, assume that in the genome there are L genes that, if mutated, could increase susceptibility to the disease. For each locus, define two classes of alleles: normal (N) and susceptibility (S) alleles. The marginal effect of each S allele is to increase the risk of disease in carriers. At any given locus, each S allele will have the same effect, but effect sizes may vary across loci.
In the subsection “Multilocus Models,” it will be necessary to make specific assumptions about the penetrance model. I will assume a model of multiplicative interactions between loci (Risch 1990a). Specifically, this means that if we use xl to designate a diploid genotype at locus l, then the penetrance for a multilocus genotype x1,x2,x3,…,xL can be written as a product Yx1Yx2Yx3…YxL. Here, Yxl is a “penetrance factor” that corresponds to genotype xl. I will assume an additive model of gene effects within loci: at each locus l, we have YSSl=YSNl+δl=YNNl+2δl, where Y..l refers to the penetrance factor for one of the three possible genotypes at locus l, and δl is a constant that depends on the locus.
It will be assumed that normal alleles mutate to susceptibility alleles at a rate μS. In practice, there typically will be many possible mutations that could impair the function of a gene. For simplicity, I treat all such mutations at a particular locus as being functionally equivalent, assuming that they lead to alleles that increase susceptibility by the same amount. Mutation can also repair susceptibility alleles, either by back mutation to the original sequence, or by compensatory mutations that repair function. Let μN be the rate of mutation from S alleles to N alleles; we might expect that typically μS≫μN, since there will be many ways to damage gene function, but fewer ways to repair any particular damage (either by an exact back mutation or by compensatory mutation). In this article, I consider both the overall frequency of S alleles and the frequencies of independent mutations to the susceptible class.
To allow for the possibility of selection acting against susceptibility alleles, it will be assumed that the relative reproductive rates of individuals who are SS, SN, or NN at a given locus are 1, 1+s, or 1+2s, respectively (s⩾0). It will be assumed that s is constant over time. This models selection as acting independently at each locus, at constant strength, and does not consider the impact of interaction among loci.
To model the frequency spectrum of disease mutations, it is also necessary to specify a demographic model for human history. Although there is still considerable uncertainty about suitable models for this, recent data from a large number of nuclear loci suggest that the frequency spectrum for human DNA sequence variation produces an acceptable fit to a model of constant population size (Cargill et al. 1999; Halushka et al. 1999). There is little evidence at autosomal loci for a systematic departure from the model of constant population size (Wall and Przeworski 2000).
Hence, in this study, I assume a model of random mating in a single population of constant effective size Ne (i.e., the standard Wright-Fisher model). On the basis of estimates of nucleotide diversity at four-fold degenerate sites (Li and Sadler 1991; Cargill et al. 1999) and of mutation rates (Giannelli et al. 1999; Nachman and Crowell 2000), the effective population size of humans (Ne) is ∼10,000 individuals.
In population genetic modeling, it is conventional to rescale the mutation and selection parameters by a factor 4Ne, because the amount and distribution of genetic variation depends only on these rescaled parameters, and it is these scaled parameters that are most easily estimated from polymorphism data (Ewens 1979; Long and Langley 1999). Thus, results in this article will be presented in terms of the scaled mutation rates βS=4NeμS and βN=4NeμN and of the selection rate σ=4Nes. The numerical values of βS and βN for typical genes are not known precisely, but rough calculations (see below) suggest that likely values for βS are in the range 0.1–1.0 and perhaps are as high as 5 at some loci. Values for βN are likely to be in the range 0.001–0.01. In view of the uncertainties in the mutation and selection parameters, results will be shown for a range of plausible values.
Clearly, the proposed model does not capture all the intricacies of reality—in part, because our understanding of the genetics of complex diseases is still in its infancy. The genetic model is very simple, with only two classes of alleles and with a very simple model of interactions between alleles and between loci. I also do not consider the impact of intralocus recombination or linkage among loci. I have estimated mutation rates for “typical” genes, but this is not intended to exclude the possibility that there will be important outliers. All of the analysis assumes that the evolutionary process is at equilibrium. The demographic model of constant population size and random mating is clearly imperfect, but it appears to provide an adequate model for the neutral frequency distribution. In this study, we need to be able to model the frequency distribution at nearly neutral loci, and so it will be assumed that this demographic model can similarly provide an adequate description for these.
Finally, it is worth discussing the implications of the assumed genetic model. Although it is often claimed that mutations for late-onset diseases may be neutral, it is unclear what sort of model to assume in testing the plausibility of this hypothesis. Here I assume two classes of alleles, and, because of the assumed asymmetric mutation, it will be shown that the S allele is usually near fixation. A related model would allow K possible levels of fitness. Limited experimentation with that model suggests that, in the neutral case, the results are similar—again, provided that mutation rates are asymmetric. Another possibility would be to allow each successive mutation to increase disease susceptibility, without limit. In the neutral case, this seems biologically implausible. It implies that the performance of the gene would deteriorate steadily because of mutation while incurring no fitness cost. But a different type of alternative may be plausible for some loci. It is possible that selection at some loci has weakened in the recent evolutionary history of humans, but mutations at these loci still increase disease susceptibility. Such a model could lead to neutral disease mutations at intermediate frequencies.
Methods
Frequency of Susceptibility Alleles
Under the model of additive selection outlined in “Models and Assumptions,” the stationary probability distribution, f(p) of the overall frequency, p, of susceptibility alleles in the population is given by Wright’s formula (Wright 1949; Ewens 1979):
where the normalization constant k can be obtained by numerical integration. (The results of the present study might be extended to more-complicated models of selection and mutation using a simulation technique developed by Fearnhead [in press].)
Simulation of Multilocus Model
The results in the section “Multilocus Models” make use of the following model. The goal is to explore the properties of a full evolutionary model of a multilocus disease while recognizing that the model contains a number of important simplifying assumptions.
Consider a disease with population prevalence K and sibling recurrence risk Λs, and assume that there are L loci that contribute to susceptibility. Suppose that susceptibility alleles at locus i increase risk by adding δi to the penetrance. Then the (additive) genetic variance caused by locus i is 2pi(1-pi)δ2i (James 1971), where pi is the frequency of susceptibility alleles at locus i. Assuming a multiplicative model of gene interactions (defined above), we have
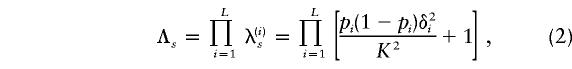 |
where λ(i)s is the contribution of locus i to the recurrence risk (Risch 1990a).
If we specify mutation and selection rates βN,i, βS,i, and σi for locus i, then the unconditional density f(pi) is given by (1). In practice, it seems that all of the underlying parameters (σ, βS, βN, and δ) will vary across loci, according to the different size, structure, and functional role of the genes. For this reason, I will assume that the parameter values at each locus are drawn independently from a set of underlying distributions.
On the basis of the arguments given previously, we might expect that βS will usually lie in the range 0.1–3.0, and βN in the range 0.001–0.01. Their distributions are unknown, but, for illustrative purposes, I will assume that the value of ln(βS) is uniformly distributed in [ln(0.1)–ln(3.0)], and the value of ln(βN) is uniformly distributed in [ln(0.001)–ln(0.01)]. These distributions place greater weight on low values: the means are 0.85 and 0.004, respectively. The distribution of σ is assumed to be the following: with probability 0.5, σ=0; otherwise σ is uniformly distributed in 0–20.0. This allows for a mixture of neutral and mildly deleterious mutations. It will be assumed that the distribution of δi is proportional to an exponential distribution with mean 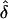 , but restricted to the range (0, 0.2). Several different values of
, but restricted to the range (0, 0.2). Several different values of 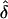 were used for the examples.
were used for the examples.
A Markov chain–Monte Carlo (MCMC) program was used to simulate from the joint posterior distribution of pi, βS,i, βN,i, σi, and δi, given K and L or given Λs, K, and L. Details are given in Appendix A.
Simulation of Genealogies with Selection
The results in the section “Frequencies and Ages of Mutations” were obtained by simulating samples of 1,000 chromosomes, using the ancestral selection graph (ASG) (Neuhauser and Krone 1997; Przeworski et al. 1999). The ASG is a recent extension of coalescent methods (Hudson 1990) to accommodate weak selection. This approach makes it possible to simulate the ancestral genealogy of a sample of chromosomes, taking into account the joint effects of mutation, selection, and genetic drift. Each sample represents an independent realization from the stochastic evolutionary process, and we can extract all the data of interest from the ancestral genealogy—in particular, the ages and frequencies of distinct mutations to the susceptible type. Details are provided in Appendix B.
Program Availability
The programs used for the computations in this article are available at my Web page.
Mutation-Rate Estimates
The extent of genetic variation at fourfold degenerate sites in humans (Li and Sadler 1991; Cargill et al. 1999) indicates that the value of 4Neμ per nucleotide site is roughly 1.0×10-3. Thus, if there were 10 or 100 sites per gene that could mutate to produce susceptibility loci, βS would be 0.01 or 0.1, respectively. The average coding length of genes in humans is ∼1,500 bp (Eyre-Walker and Keightley 1999). If every nonsynonymous mutation (∼3/4 of all possible mutations) produced a susceptibility allele, the average for βS would be ∼1.1 (and possibly a little higher, if we consider noncoding regulatory sites and length variants). In practice, however, only some fraction of nonsynonymous mutations—that is, those that change functionally critical amino acids—will lead to alleles with measurable phenotypic effects. For example, at the hemophilia B locus, it has been estimated that the target region for detrimental mutations is 275 nucleotides, out of a total length of 1,362 (Giannelli et al. 1999). A similar result is suggested indirectly by Eyre-Walker and Keightley (1999), who, using divergence data from a number of genes, estimated that, on average, 38% of nonsynonymous mutations in humans are eliminated by natural selection. On the basis of these arguments, a plausible estimate for typical values of βS might be in the range 0.1–1.0.
Direct estimates of the mutation rate for a number of Mendelian disorders are generally consistent with this, or slightly higher. For instance, mutation rates per generation have been estimated for neurofibromatosis (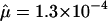 to 4.3×10-5, Friedman 1999), spinal muscular atrophy (
to 4.3×10-5, Friedman 1999), spinal muscular atrophy (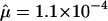 , Wirth et al. 1997), hemophilia B (
, Wirth et al. 1997), hemophilia B (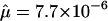 , Green et al. 1999), and Apert syndrome (
, Green et al. 1999), and Apert syndrome (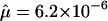 , Tolarova et al. 1997). Taking Ne=10,000, these correspond to scaled mutation rates of 4Neμ=1.7 to 5.1, 4.4, 0.3, and 0.2, respectively. However, it is not clear that mutation rates for Mendelian disorders will necessarily be representative of complex-disease loci. There may also be a publication bias toward loci with high mutation rates, since the corresponding diseases will be more common at mutation-selection balance. Nonetheless, in this article, I present results for βS in the range 0.1–5.0.
, Tolarova et al. 1997). Taking Ne=10,000, these correspond to scaled mutation rates of 4Neμ=1.7 to 5.1, 4.4, 0.3, and 0.2, respectively. However, it is not clear that mutation rates for Mendelian disorders will necessarily be representative of complex-disease loci. There may also be a publication bias toward loci with high mutation rates, since the corresponding diseases will be more common at mutation-selection balance. Nonetheless, in this article, I present results for βS in the range 0.1–5.0.
A standard argument holds that the repair rate βN will typically be much smaller than βS, because there may be many ways to impair the function of a gene, but mutations that repair the damage must be rather specific. Repair would proceed either by an exact reversion, or by compensatory mutation at a limited number of other sites. There is strong support for this qualitative argument in model organisms—for instance, in experiments showing the action of Muller’s ratchet in highly inbred populations (e.g., Chao 1990) and in various studies of mutation accumulation. Similarly, a recent study in Caenorhabditis elegans found that compensatory mutations had no measurable effect in ameliorating the mutation load in mutagenized lines (Peters and Keightley 2000). However, data estimating the actual rates of compensatory mutation are sparse. One exception is a study of reversion to streptomycin resistance in Escherichia coli, which found that a small number of nonsilent mutations occurred repeatedly in 24 independent lines, implying a limited number of possible compensatory mutations—the authors suggest 10–20 (Levin et al. 2000). Since βN appears to be substantially smaller than βS, but must be at least as large as the rate of back mutation, an estimate in the range 0.0003–0.01 would seem reasonable for typical values of βN. This would correspond to there being 1–30 possible compensatory mutations per gene (each mutation occurs at 1/3 of the total rate per site), consistent with Levin et al. (2000).
Results
Overall Frequency of Susceptibility Alleles at a Disease Locus
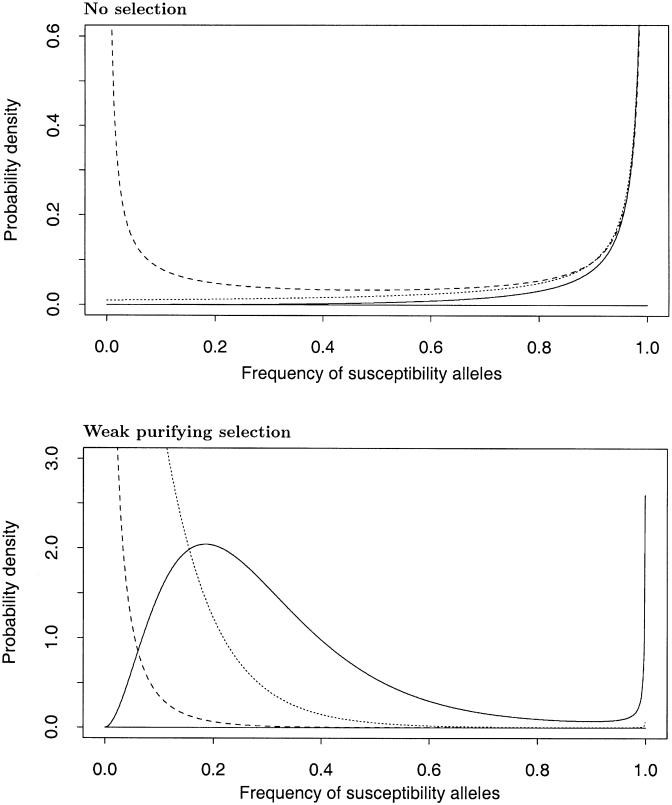
At any given locus, the frequency of susceptibility alleles is a random quantity resulting from the joint effects of selection, mutation, and random genetic drift. The stationary probability distribution, f(p), for the overall frequency, p, of susceptibility alleles under the assumed model is given by equation (1). Examples of plots of this probability distribution are shown in figure 1. Except at very high mutation rates, these plots exhibit a characteristic U-shaped distribution, in which much of the probability density is on p near 0 or 1.
Figure 1.
Examples of the probability distribution of the overall frequency of susceptibility alleles at a locus (from Wright’s formula). In the upper plot, σ=0; in the lower plot, σ=12.0. Parameter values: βS=3.0 (solid lines); βS=1.0 (dotted lines); βS=0.1 (dashed lines); βN=0.01 throughout. Notice that the vertical scale differs by a factor of five between the plots. In the upper plot, virtually all of the probability mass is on values near 0 or 1 (see table 1).
Figure 1A shows distributions of p, in the absence of selection, for a range of mutation rates. As outlined above, we might expect that, at most loci, βS≫βN, with typical values for βS being in the range 0.1–5.0 and for βN in the range 0.001–0.01. At these mutation rates, the susceptible type is highly unlikely to be at intermediate frequencies in the absence of selection (see also table 1A). It is much more likely to be near fixation (in which case, we might describe the N allele as being “protective,” rather than normal). Only if βN is quite large (e.g., ⩾0.1) and of the same magnitude as βS, is there an appreciable probability that the susceptible type will be at intermediate frequencies (table 1A).
Table 1.
Probability that a Locus is Polymorphic for Susceptibility Alleles[Note]
|
βN |
||||
| βS | .001 | .01 | .1 | 1.0 |
| A. No Selection |
||||
| .001 | .0046 | .0082 | .0080 | .0046 |
| .01 | .0082 | .0448 | .0729 | .0449 |
| .1 | .0080 | .0729 | .3593 | .3680 |
| .5 | .0058 | .0559 | .4247 | .8950 |
| 1.0 | .0046 | .0449 | .3680 | .9800 |
| 5.0 | .0026 | .0254 | .2309 | .9510 |
| 10.0 | .0019 | .0185 | .1755 | .9043 |
| B. Weak Purifying Selection (σ=12) |
||||
| .001 | .0017 | .0017 | .0017 | .0017 |
| .01 | .0173 | .0173 | .0173 | .0166 |
| .1 | .1656 | .1656 | .1650 | .1587 |
| .5 | .6332 | .6396 | .6387 | .6242 |
| 1.0 | .8410 | .8912 | .8956 | .8869 |
| 5.0 | .0319 | .2534 | .8241 | .9993 |
| 10.0 | .0049 | .0476 | .3781 | .9859 |
Note.— Probability that a locus is polymorphic for susceptibility alleles (i.e., that the frequency of S lies between .01 and .99); βS and βN give the scaled mutation rates to and from the susceptible class of alleles. Rough calculations suggest that, for most genes, βS will be in the range 0.1–5.0, and we can expect that the “repair” rate βN will typically be rather smaller: probably of the order of ⩽0.01.
Figure 1B illustrates the effect of selection against susceptibility alleles. Crucially, weak purifying selection against S alleles can have the effect of greatly increasing the probability that the susceptible type will be polymorphic at any given locus, even when the “repair” rate βN is small (see also table 1B). This effect is especially important when the S allele suffers only a very small selective disadvantage. For instance, the selection rate shown in the figure (σ=12) corresponds to a selective disadvantage for S alleles of ∼3×10-4 per generation.
The reason that weak purifying selection increases polymorphism is that it greatly reduces the probability that susceptibility alleles will be at or near fixation, but it is not so strong that it prevents S alleles from reaching intermediate frequencies. The situation is different when the susceptibility alleles are very deleterious—as seen at Mendelian disease loci—in which case, selection dominates the effects of mutation pressure and drift and keeps S alleles at low frequency.
Contribution to Genetic Variance as a Function of Allele Frequency
Another way to characterize the frequency distribution is in terms of contribution to genetic variance for the trait of interest. In view of the property that the distribution of S-allele frequencies tends to be U shaped, it is natural to ask whether the bulk of the genetic variance is due to a small number of loci where susceptibility alleles are common or is due to a much larger number of loci where susceptibility alleles are quite rare. In this section, I consider the marginal distribution at one locus; in the next section, I describe results under a full multilocus model.
For concreteness, consider a locus at which the marginal effect of each susceptibility allele is to add δ to the penetrance (see “Models and Assumptions”). Recall that the genetic variance is given by 2p(1-p)δ2, where p is the frequency of susceptibility alleles. Integrating over allele frequencies, the expectation of the additive genetic variance caused by this locus is
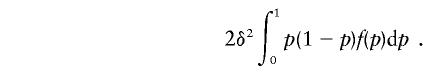 |
If we have a large number of such loci, the contribution of loci with an allele frequency p to the additive genetic variance is proportional to p(1-p)f(p).
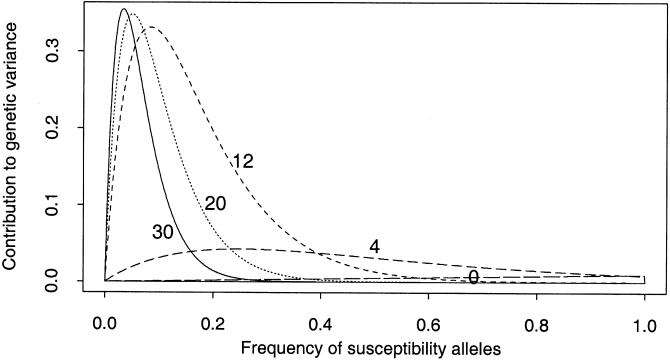
Figure 2 shows examples of plots of p(1-p)f(p), for plausible mutation rates, and a range of values of the selection coefficient. With weak purifying selection, most of the contribution to genetic variance comes from low or intermediate allele frequencies (e.g., <0.3). In the absence of selection, medium or high frequencies contribute much of the genetic variance, but, in this case, the total expected genetic variance (proportional to the integrals of these curves) is very low indeed.
Figure 2.
Contribution to the additive genetic variance as a function of allele frequency. The plot shows how much of the (expected) additive genetic variance is due to alleles of a given frequency. The vertical axis is in units of 2δ2, where δ is the marginal increase in penetrance caused by each susceptibility allele. The integral of each curve equals the expected additive genetic variance. The lines are labeled with the assumed values of σ: 0.0, 4.0, 12.0, 20.0, and 30.0; mutation rates are βS=1.0, βN=0.01 for all.
The results shown so far suggest a number of features of the proposed model. First, the great majority of potential disease-susceptibility loci will have essentially no genetic variation (and contribute little to variance in phenotype), unless (1) the S alleles are under weak purifying selection or (2) the repair rate βN is surprisingly high. In the presence of purifying selection, loci with high forward mutation rates βS tend to be more variable and contribute more to genetic variance than do loci with low βS. Notice that these predictions depend on the underlying population genetic model, but (unlike in the following section) do not depend much on the specifics of the penetrance model.
Multilocus Models
So far, I have described the marginal properties of a single locus under the proposed model. I now present results from Monte Carlo simulation of the full multilocus model. In brief, the elements of that model are that L loci interact multiplicatively to produce a penetrance for the phenotype of interest. The frequency of susceptibility alleles at each locus is a random variable, the distribution of which depends on the population parameters σ, βS, and βN. The values of these parameters (and also of δ) are assumed to vary across loci. To model the variation in parameters across loci, the values at each locus are assumed to be drawn from some underlying distributions. These distributions are, of course, unknown, and so the results presented assume a particular choice of plausible distributions. The models and assumptions used in this section of the article are less general and more speculative than in the other sections of this article. As such, the results should be treated as exploratory.
Table 2 shows expected values of λs for the major disease loci under a range of disease scenarios. Values of λs are important determinants of the power of affected sib-pair studies (Risch 1990b). It is interesting that for many of the parameter sets shown here, a single locus is responsible for much of the total recurrence risk Λs. In such situations, it would be relatively easy to map the major locus using affected sib pairs, suggesting that these parameter sets may not be representative of most complex diseases, where experience shows that high LOD scores are unusual.
Table 2.
Expected Values of the Single-Locus Contributions (λs) to Recurrence-Risk Ratio, under the Multilocus Model Described in the Text[Note]
|
Average Recurrence-Risk Ratios |
|||||||
ModelParameters(K, Λs, L, 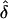 ) ) |
λ[1]s | λ[2]s | λ[3]s | λ[4]s | λ[5]s | i=6Lλ[i]s | F(Λs) |
| .05, 5, 5, .10 | 3.28 | 1.54 | 1.05 | 1.00 | 1.00 | … | .99 |
| .05, 10, 20, .10 | 2.94 | 1.96 | 1.43 | 1.17 | 1.06 | 1.04 | .96 |
| .05, 10, 100, .10 | 1.92 | 1.52 | 1.34 | 1.24 | 1.18 | 1.85 | .05 |
| .01, 10, 5, .02 | 7.90 | 1.37 | 1.04 | 1.00 | 1.00 | … | .98 |
| .01, 10, 20, .02 | 4.10 | 1.80 | 1.29 | 1.12 | 1.05 | 1.03 | .75 |
| .01, 10, 50, .02 | 2.70 | 1.65 | 1.36 | 1.22 | 1.14 | 1.50 | .20 |
| .01, 10, 70, .02 | 2.41 | 1.61 | 1.35 | 1.23 | 1.16 | 1.46 | .05 |
| .001, 10, 5, .005 | 7.38 | 1.49 | 1.04 | 1.00 | 1.00 | … | .81 |
| .001, 10, 20, .005 | 4.08 | 1.81 | 1.30 | 1.12 | 1.05 | 1.02 | .23 |
| .001, 50, 20, .005 | 8.16 | 3.00 | 1.72 | 1.28 | 1.12 | 1.08 | .45 |
| .0004, 75, 20, .001 | 8.33 | 3.47 | 1.90 | 1.39 | 1.17 | 1.05 | .86 |
| .0004, 75, 100, .0005 | 3.89 | 2.20 | 1.75 | 1.50 | 1.34 | 3.09 | .60 |
Note.— The values of λ[i]s are ordered by size; for instance, the column λ[1]s gives the expected value of λs at the locus that makes the largest contribution to Λs. The column λ[i]s gives the expected value of the product of the L-5 smallest values of λs. F(Λs) gives the probability that in unconditional simulations with these parameter values, the sibling recurrence-risk ratio is less than the assumed Λs. Parameters: K, disease prevalence; Λs, sibling recurrence-risk ratio; L, number of loci; 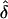 , shape parameter for the distribution of allele effect sizes (approximately the mean of δ). The products of the row values do not equal Λs because the λ[i]s are arithmetic means.
, shape parameter for the distribution of allele effect sizes (approximately the mean of δ). The products of the row values do not equal Λs because the λ[i]s are arithmetic means.
In contrast, in those cases where L is large (and also if 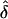 is quite small compared to K), the distribution of λs is flatter. The parameter values used for the last two lines of table 2 are loosely based on a study of autism (Risch et al. 1999). That article described a genome screen of autistic sib pairs in which there were no strong linkage results, and most of the genome was excluded from containing loci with λs⩾3.0. The authors argued that in view of the high heritability of autism (Λs≈75), this must indicate a large number of genes of modest effect. Assuming a multiplicative model with L loci with equal values of λs, they calculated that there must be ∼15 genes or more. But the results here suggest that the distribution of λs is likely to be quite skewed, and so a model with as few as 20 loci does not fit their findings very well. A model with 100 loci (and correspondingly smaller
is quite small compared to K), the distribution of λs is flatter. The parameter values used for the last two lines of table 2 are loosely based on a study of autism (Risch et al. 1999). That article described a genome screen of autistic sib pairs in which there were no strong linkage results, and most of the genome was excluded from containing loci with λs⩾3.0. The authors argued that in view of the high heritability of autism (Λs≈75), this must indicate a large number of genes of modest effect. Assuming a multiplicative model with L loci with equal values of λs, they calculated that there must be ∼15 genes or more. But the results here suggest that the distribution of λs is likely to be quite skewed, and so a model with as few as 20 loci does not fit their findings very well. A model with 100 loci (and correspondingly smaller 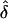 ) fits rather better. In this case, the expected λs for the top 15 loci is 3.89–1.04, whereas the remainder of the loci make virtually no contribution to the population risk.
) fits rather better. In this case, the expected λs for the top 15 loci is 3.89–1.04, whereas the remainder of the loci make virtually no contribution to the population risk.
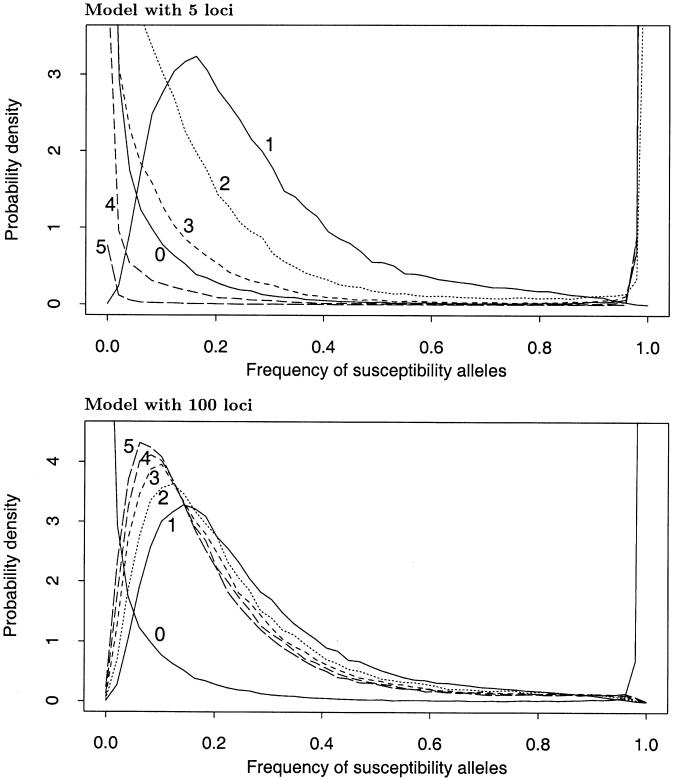
Figure 3 plots the allele frequency distributions at the top five loci under two of the models from table 2. In both cases, the allele frequencies at loci that contribute substantially to Λs are shifted away from the null distribution and toward intermediate frequencies.
Figure 3.
Probability distributions of the overall frequency of susceptibility alleles at each locus, under the multilocus model described in the text. In each plot, the solid line labeled “0” shows the unconditional distribution of allele frequencies. The lines labeled “1”,…,“5” show the frequency distributions at the five loci that contribute the most to the sibling recurrence risk: that is, line “i” gives the frequency distribution at the locus with the ith largest value of λs. Parameters: (top) K=0.01, Λs=10.0, L=5, and 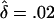 ; (bottom) K=0.0004, Λs=75.0, L=100, and
; (bottom) K=0.0004, Λs=75.0, L=100, and 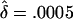 .
.
Finally, table 3 shows the expected values of the various parameters under three different disease scenarios. Notice that the loci with large values of λs have high average values of σ, βS, and (usually) δ, compared with loci that have low values of λs and with the unconditional distributions. This observation reflects the property, described above, that loci with purifying selection and high mutation rates are more likely to be genetically variable and, hence, contribute more to variance in disease susceptibility. Recall that δ and σ were modeled as independent. In practice, we might expect them to be positively correlated, which should have the effect of strengthening this result.
Table 3.
Expected Values of Key Parameters (Selection, σ; Mutation, βS,βN; and Allele-Effect Size, δ) under a Range of Disease Scenarios, under the Multilocus Model and Prior Distributions Described in the Text[Note]
|
λ[i]s |
||||||
| K,Λs,Land KeyParameters | 1 | 2 | 3 | 4 | 5 | 20 |
| .01,10,5: | ||||||
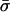
|
11.1 | 10.7 | 6.75 | 2.95 | 1.32 | … |
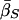
|
1.30 | 1.00 | .85 | .86 | .91 | … |
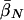
|
.004 | .004 | .004 | .004 | .004 | … |
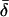
|
.067 | .018 | .013 | .013 | .013 | … |
| .01,10,20: | ||||||
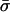
|
11.5 | 11.9 | 12.0 | 11.9 | 11.3 | 1.04 |
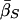
|
1.25 | 1.16 | 1.08 | 1.00 | .91 | .93 |
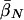
|
.004 | .004 | .004 | .004 | .004 | .004 |
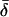
|
.048 | .029 | .019 | .014 | .012 | .016 |
| .001,10,5: | ||||||
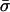
|
12.3 | 8.73 | 3.93 | 1.66 | 1.08 | … |
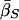
|
1.01 | .81 | .82 | .90 | .92 | … |
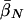
|
.004 | .004 | .005 | .005 | .004 | … |
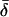
|
.012 | .009 | .012 | .013 | .014 | … |
| Unconditional: | ||||||
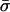
|
5.76 | |||||
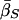
|
.84 | |||||
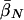
|
.004 | |||||
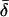
|
.020 | |||||
Note.— The data in the table give the expected values for each parameter, ordered by λ[i]s. That is, the column of data marked “i” gives the expected values of σ, etc., at the locus with the ith largest value of λs. Notice that the loci that make the largest contribution tend to have stronger selection and higher mutation rates to susceptibility alleles than average. The “unconditional” results show the expectations of the parameters under the assumed prior distributions (see Methods and Appendix). 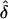 was 0.02 in all four examples.
was 0.02 in all four examples.
It should be noted that the strongly skewed distributions of λs may be due, in part, to the choice of a multiplicative model of gene interactions. In this model, it is very easy for one locus or a few loci to contribute most of Λ (purely because the λs are multiplied to produce Λ). Other models of gene interaction—for instance, models with heterogeneity—may lead to less skewed distributions of λs. These are technically more difficult to analyze, but this analysis will probably be a useful task for the future. The other results from this section are similar to those obtained under the single-locus model above, and so it seems likely that they will be robust to changes in the model of gene interactions.
Frequencies and Ages of Mutations
So far, I have considered the total frequency of susceptibility alleles. This frequency might include contributions from several independent mutations. For association or linkage-disequilibrium mapping, it is most important to know about the frequencies and ages of individual mutations within the susceptible class, since each new mutation (usually) occurs on a different haplotype background. If there are multiple mutations, no single haplotype will be strongly associated with the disease. In the context of models such as that of Risch and Merikangas (1996), this reduces the genotypic risk factor associated with any particular mutation, thus lowering the power of standard tests of association (e.g., Spielman et al. 1993).
To study the frequencies of individual mutations, I have conducted simulations of the evolutionary process at a disease locus, incorporating mutation, selection, and drift (see Methods). Table 4 shows a summary of results for three different mutation rates to the susceptible type. At a low mutation rate (βS=0.1), it is usually the case that most S alleles are descended from a single disease mutation (see “Fraction” in table 4). However, when the mutation rate is high (βS=5.0), it is unlikely that any particular mutation is at high frequency within the susceptible class, making association mapping much more difficult. This result does not depend strongly on the overall frequency of S alleles, on the repair rate βN (results not shown), or on the presence or absence of weak selection (see the results of Slatkin and Rannala [1997], who used a similar mutation model).
Table 4.
Properties of the Most Common Susceptibility Mutation, as a Function of the Mutation Rate to the Susceptible Type[Note]
|
Neutral Mutations |
With Selection |
|||||
| βS andFrequencyof S | Fraction | Age(years × 103) | IBD(cM) | Fraction | Age(years × 103) | IBD(cM) |
| .1: | ||||||
| 1%–5% | .94 | 70 | .23 | .94 | 37 | .28 |
| 5%–10% | .94 | 156 | .08 | .94 | 74 | .11 |
| 10%–20% | .94 | 244 | .04 | .94 | 103 | .07 |
| 20%–50% | .95 | 403 | .02 | .94 | 150 | .04 |
| 1.0: | ||||||
| 1%–5% | .64 | 52 | .27 | .64 | 33 | .33 |
| 5%–10% | .65 | 110 | .12 | .65 | 61 | .14 |
| 10%–20% | .66 | 164 | .06 | .65 | 91 | .08 |
| 20%–50% | .70 | 288 | .03 | .67 | 136 | .05 |
| 5.0: | ||||||
| 1%–5% | … | … | … | .32 | 24 | .48 |
| 5%–10% | … | … | … | .33 | 38 | .25 |
| 10%–20% | .31 | 66 | .13 | .34 | 56 | .14 |
| 20%–50% | .42 | 169 | .04 | .36 | 84 | .08 |
Note.— Results are binned according to the overall frequency of S alleles. “Fraction” is the average frequency of the most common mutation among all S alleles; “Age” is the average age of the most-common mutation (Ne=10,000 and generation time = 20 years); “IBD” gives the average length (in cM) that is shared between two chromosomes carrying the most common mutation. Two rows are left blank because these frequencies of S alleles occurred too rarely in simulations to allow accurate estimates. Parameter values: βN=0.01 for all; left column: σ=0; right column: σ=12 (upper two blocks), σ=20 (lower block). Results include only those realizations in which the sample MRCA is of type N.
Note that, in Mendelian disorders, strong selection ensures that no single mutation reaches high frequencies, with the effect that the susceptible class is usually made up of many distinct mutations, all at low frequency (Slatkin and Rannala 1997) as is seen in practice (e.g., Green et al. 1999).
Table 4 also shows the ages of mutations and the average length of haplotype that is shared between two chromosomes carrying the most common mutation. The length of shared haplotype (and, hence, the region in which association tests are potentially powerful) increases substantially when the overall frequency of S alleles is low. The length of shared haplotype is also larger when there is weak selection than in the neutral case (since the mutations tend to be younger).
It is interesting to consider the results from this section in light of those from the previous sections. In particular, susceptibility loci with large values of λs—that is, the loci that we have the best chance of finding by linkage methods—can be expected to have relatively high mutation rates (βS), perhaps of the order of ⩾1.0, and are probably at low-to-intermediate frequencies. High mutation rates are unfortunate from the point of view of linkage-disequilibrium mapping, because they imply that there will often be significant allelic heterogeneity. However, table 4 does imply that we might expect relatively large regions of identical-by-descent (IBD) sharing around disease mutations. For instance, this might often be of the order of 0.1 cM (∼100 kb, on average).
Discussion
This study models the genetic variation at disease-susceptibility loci, taking into account the evolutionary processes, including mutation, genetic drift, and the possibility of selection. The values of the parameters underlying the proposed model—particularly the mutation rates to and from the susceptible type—are not well characterized; however, it is possible to estimate likely values.
On the basis of this model, I have examined several issues of practical importance for gene mapping. The first of these is the overall frequency of susceptibility mutations. If we assume a neutral model, then, for the most plausible mutation rates, the probability that susceptibility alleles are at intermediate frequencies at any given locus is rather low. This is because the mutation rate βS is expected, on biological grounds, to be much higher than the repair rate βN. Hence, in the absence of selection, S alleles will be near fixation at most susceptibility loci. Loci that are fixed for S alleles contribute to genetic risk, but, because they are not variable, they do not contribute to differences among individuals and cannot be mapped by conventional methods. Mutations with exclusively late-onset effects could fall into this category. By contrast, loci at which there is weak purifying selection against susceptibility alleles are much more likely to be variable, because selection makes it improbable that S alleles reach high frequency.
Another way to look at this is in terms of the expected contribution that a susceptibility locus makes to the genetic variance underlying a trait. Unless the repair rate βN is very high, neutral loci contribute very little to the genetic variance. We can expect that, in practice, the mutation rate and strength of selection will vary across loci. Although the joint distribution of these is not known, the results presented here indicate that loci with large values of βS and weak purifying selection will tend to contribute disproportionately to the genetic variance. Even very weak selection (of the order of 10-4 per generation) has a significant purifying effect. These results are predicted both from the (relatively simple) single-locus model presented and from the full multilocus model.
I have also modeled the number of distinct mutations that contribute to the susceptible class at any given locus (i.e., allelic heterogeneity). The predicted range of mutation rates (βS) covers a critical region. At loci where βS is low, the susceptible class will usually be dominated by a single major mutation. But if βS is in the upper end of the predicted range, then it is unlikely that any single mutation will constitute a large fraction of the susceptible class. In this case, association mapping is not very powerful (Slager et al. 2000). As noted above, loci with high mutation rates contribute disproportionately to the genetic variance and to Λs, and these are the loci that will most easily be mapped by linkage methods. Hence, allelic heterogeneity is likely to pose a severe challenge for fine mapping.
The title of this article asks whether rare variants are responsible for susceptibility to complex diseases (the reverse of the “common disease–common variant” hypothesis). From the results shown here, there are different answers to this, depending on what exactly is meant by the question. If we select a random susceptibility locus, S alleles are quite likely to be at a frequency near 0 or 1 (fig. 1). But for any given disease, the loci that contribute substantially to the genetic variance, or to Λs, are more polymorphic than random loci. At such loci, the frequency of susceptibility alleles ranges from rare (e.g., 1%) to quite common (e.g., 50%) for most of the parameter values shown in figures 2 and 3. What about the frequencies of the individual variants (i.e., distinct mutations to the S class)? The loci with the largest values of λs are likely to have high mutation rates; results shown in table 4 suggest that at such loci, the most common variant has an expected frequency of about half the total frequency of S alleles.
In summary, the findings have a number of implications for gene mapping:
-
(1)
Many loci that are good biological candidates for contributing to a particular disease will not be polymorphic for susceptibility alleles, simply as a result of the randomness of the evolutionary processes (table 1).
-
(2)
It is critical to develop statistical methods for testing for association that are powerful in the presence of allelic heterogeneity. Instead of testing for association at just one or a few SNPs at a time, these might proceed by identifying sets of haplotypes that appear with high frequencies in affected individuals.
-
(3)
The results suggest that most susceptibility mutations that are polymorphic will be mildly deleterious. If this is the case, then susceptibility mutations will usually be at sites that are selectively constrained and will therefore have low rates of evolutionary divergence between species. Thus, sequence comparisons between species should be a useful tool for interpretation of genetic variation in putative disease genes and for identification of functional sites.
-
(4)
In addition to allelic heterogeneity, power calculations on the efficiency of linkage-disequilibrium mapping (e.g., Kruglyak 1999) need to consider mutations at low frequency and to incorporate weak selection. Both of these effects substantially increase the extent of haplotype sharing around a disease mutation. Empirical studies of the extent of linkage disequilibrium (e.g., Taillon-Miller et al. 2000) should pay particular attention to the properties of low-frequency SNPs.
-
(5)
The results obtained here provide mixed support for the contention that association mapping will be more powerful than family-based methods for finding complex-disease genes (Risch and Merikangas 1996). The advantage of association mapping, compared with linkage methods, is particularly large when the susceptibility allele is rare (Risch and Merikangas 1996); it is also encouraging for the linkage-disequilibrium approach that regions of haplotype sharing are predicted to be rather large. However, allelic heterogeneity will considerably reduce the power of association methods (but not of family-based methods) until appropriate statistical techniques are developed.
Acknowledgments
This work was supported by a Hitchings-Elion grant from the Burroughs-Wellcome Fund. I thank P. Donnelly, P. Fearnhead, G. McVean, M. Przeworski, and M. Stephens for helpful comments and discussions. I also thank the reviewers for their careful reading of the manuscript and their thoughtful comments.
Appendix A: MCMC Algorithm
The MCMC algorithm used for the section “Multilocus Models” was as follows. The version that conditions on Λs is described as “conditional.” For background on MCMC methods, see Gilks et al. (1996).
For each locus i, arbitrary initial values of βS,i, βN,i, and σi were chosen (e.g., by making independent draws from the priors). The δi and pi were also chosen randomly in the unconditional case, but, in the conditional case, they were chosen in such a way as to produce the correct value of Λs (eq. 2). The following steps were then iterated.
-
1. For each i, proposal values of β′S,i, β′N,i, and σ′i were simulated from their respective prior distributions. The new values were accepted with probability
where f(p) is given by (1). Otherwise, the old values were retained. In practice, each of βS, βN, and σ were usually updated in separate Metropolis-Hastings steps.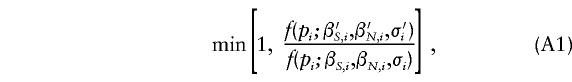
-
2a.(Not conditional on Λs.) For each locus i, a new value of δi was simulated from the assumed prior distribution and was accepted with probability 1. One of two proposal densities was used to update pi. With some probability (e.g., 0.5), p′i was Uniform(0,1), in which case it was accepted with probability
Otherwise, p′i was Beta(βS,i,βN,i), in which case it was accepted with probability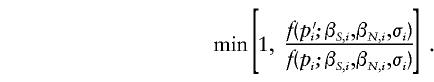

-
2b.(Conditional on Λs.) We need to construct moves that update the pi and δi without changing Λs. This was done by updating one pi and one δj simultaneously (where i and j denote loci, and i may equal j). The proposal p′i was chosen from one of the two proposal distributions used in (2a), and then δ′j was chosen so that Λs remained unchanged. When this was being done, δ′j might be outside the allowed range of δ, in which case both p′i and δ′j were rejected. Otherwise, p′i and δ′j were accepted with probabilities given by
and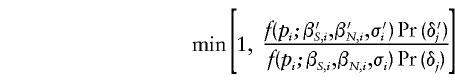
for the Uniform and Beta proposals, respectively, where Pr(δj) refers to the probability under the assumed prior distribution.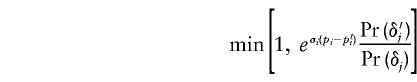
Data were collected by sampling values of pi, δi, etc., from the Markov chain, after some suitable dememorization period. In evaluating equation (A1), it is necessary to compute the normalizing constant for Wright’s formula. When σ≠0, this must be done by numerical integration. My approach here was to use MATHEMATICA to compute a table of values of the normalizing constant (for a range of values of βS, βN, and σ). I then used numerical interpolation to approximate the normalizing constants for each proposal.
For some values of the mutation and selection rates, the probability density for p (eq. 1) is very sharply spiked near 0 and 1. This means that some care must be taken in choosing the proposal distribution for pi; otherwise, the Markov chain may fail to visit these regions. The choice of proposals used here (a mixture of a Beta and a Uniform) seems to allow fairly good mixing; in contrast, a Uniform proposal used alone performs extremely poorly.
I have performed several tests of the performance of the MCMC code. Convergence appears to be fairly rapid, and independent runs produce no indication of multimodality. The distribution of pi (in the unconditional case) matches the target distribution (eq. 1) when the population parameters are fixed. However, two lines of evidence suggest that the performance is not ideal. First, in the unconditional case, the posterior distribution of all the parameters should match their priors. The first implementation of the program performed rather badly in this respect, but adjusting the algorithm (primarily the proposals for pi) has largely fixed this problem (the posterior mean for σ is still a bit high: ∼5.7 instead of 5.0). Second—again, in the unconditional case—we can record the parameter values whenever the overall Λs is near some specified value. Running the conditional version of the Markov chain at this Λs produces closely similar, but not identical, results. These small discrepancies are probably due to a combination of inaccuracy in the normalizing constants and the difficulty of exploring the very spiked density for p.
Appendix B: Ancestral Selection Graph
The ancestral selection graph simulations were run backwards in time and were stopped either when the ultimate ancestor was reached or when a large amount of coalescent time had passed. In both cases, the genotypes (N or S) at the point where the process was stopped were drawn from the joint stationary distribution given by Wright’s formula. The stopping time was chosen so that the probability of it being shorter than the most recent common ancestor (MRCA) time was miniscule (and this event never occurred during the simulations presented here). The simulations assumed genic selection, in which—moving backward in time—each lineage branches into two, at the rate of σ/2. The mutation rates to N and to S (moving forward in time) were βN/2 and βS/2, per lineage, respectively. The mutation rates within allelic classes (i.e., S→S, and N→N) were zero. In order to simplify the presentation, the results include only those realizations for which the MRCA of the sample is of type N.
The ages of mutations were converted from coalescent time to years by multiplying by 2gNe, where the generation time g was taken as 20 years and Ne was taken as 10,000. The expected size of region shared IBD, in cM, between pairs of chromosomes in each replicate simulation was estimated by computing 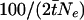 , where
, where 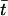 is the mean coalescent time for pairs of chromosomes carrying the most common disease mutation.
is the mean coalescent time for pairs of chromosomes carrying the most common disease mutation.
Electronic-Database Information
The URL for data in this article is as follows:
- Author's Web site, http://www.stats.ox.ac.uk/~pritch/home.html (for programs used)
References
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–238 [DOI] [PubMed] [Google Scholar]
- Chakravarti A (1999) Population genetics—making sense out of sequence. Nat Genet 21:56–60 [DOI] [PubMed] [Google Scholar]
- Chao L (1990) Fitness of RNA virus decreased by Muller’s ratchet. Nature 348:454–455 [DOI] [PubMed] [Google Scholar]
- Ewens WJ (1979) Mathematical population genetics. Springer-Verlag, New York [Google Scholar]
- Eyre-Walker A, Keightley PD (1999) High genomic deleterious mutation rates in hominids. Nature 397:344–347 [DOI] [PubMed] [Google Scholar]
- Fearnhead P (2001) Perfect simulation from population genetic models with selection. Theor Pop Biol (in press) [DOI] [PubMed] [Google Scholar]
- Friedman JM (1999) Epidemiology of neurofibromatosis type 1. Am J Med Genet 89:1–6 [PubMed] [Google Scholar]
- Giannelli F, Anagnostopoulos T, Green PM (1999) Mutation rates in humans. II. Sporadic mutation-specific rates and rate of detrimental mutations inferred from Hemophilia B. Am J Hum Genet 65:1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks WR, Richardson S, Spiegelhalter, DJ (eds) (1996) Markov Chain Monte Carlo in practice. Chapman & Hall, London [Google Scholar]
- Green PM, Saad S, Lewis CM, Giannelli F (1999) Mutation rates in humans. I. Overall and sex-specific rates obtained from a population study of hemophilia B. Am J Hum Genet 65:1572–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22:239–247 [DOI] [PubMed] [Google Scholar]
- Hudson RR (1990) Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J (eds) Oxford surveys in evolutionary biology, vol. 7. Oxford University Press, Oxford, pp 1–44 [Google Scholar]
- James JW (1971) Frequency in relatives for an all-or-none trait. Ann Hum Genet 35:47–49 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Lander ES (1996) The new genomics: global views of biology. Science 274:536–539 [DOI] [PubMed] [Google Scholar]
- Levin BR, Perrot V, Walker N (2000) Compensatory mutations, antibiotic resistance, and the population genetics of adaptive evolution in bacteria. Genetics 154:985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H, Sadler L (1991) Low nucleotide diversity in man. Genetics 129:513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Langley CH (1999) The power of association studies to detect the contribution of candidate gene loci to variation in complex traits. Genome Res 9:720–731 [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser C, Krone SK (1997) The genealogy of samples in models with selection. Genetics 145:519–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AD, Keightley PD (2000) A test for epistasis among induced mutations in Caenorhabditis elegans. Genetics 156:1635–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Charlesworth B, Wall JD (1999) Genealogies and weak purifying selection. Mol Biol Evol 16:246–252 [DOI] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, DiRienzo A (2000) Adjusting the focus on human variation. Trends Genet 16:296–302 [DOI] [PubMed] [Google Scholar]
- Risch N (1990a) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Risch N (1990b) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Risch N (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager SL, Huang J, Vieland, VJ (2000) Effect of allelic heterogeneity on the power of the transmission disequilibrium test. Genet Epidemiol 18:143–156 [DOI] [PubMed] [Google Scholar]
- Slatkin M, Rannala B (1997) The sampling distribution of disease-associated alleles. Genetics 147:1855–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–513 [PMC free article] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice J, Kwok PY (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet 25:324–328 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM (1998) Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotech 9:578–594 [DOI] [PubMed] [Google Scholar]
- Tolarova MM, Harris JA, Ordway DE, Vargerik K (1997) Birth prevalence, mutation rate, sex ratio, parents’ age, and ethnicity in Apert Syndrome. Am J Med Genet 72:394–398 [DOI] [PubMed] [Google Scholar]
- Wall JD, Przeworski M (2000) When did the human population start increasing? Genetics 155:1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, B, Schmidt T, Hahnen E, Rudnik-Schöneborn S, Krawczak M, Möller-Myhsok B, Schönling J, Zerres K (1997) De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet 61:1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S (1949) Adaptation and selection. In: Jepson G, Simpson G, Mayr E (eds) Genetics, palaeontology and evolution. Princeton University Press, Princeton, pp 365–389 [Google Scholar]
- Zöllner S, von Haeseler A (2000) A coalescent approach to study linkage disequilibrium between single-nucleotide polymorphisms. Am J Hum Genet 66:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick ME, Cutler DJ, Chakravarti A (2000) Patterns of genetic variation in Mendelian and complex traits. Annu Rev Genomics Hum Genet 1:387–407 [DOI] [PubMed] [Google Scholar]