Abstract
Zellweger syndrome (ZS), neonatal adrenoleukodystrophy (NALD), and infantile Refsum disease (IRD) are clinically overlapping syndromes, collectively called “peroxisome biogenesis disorders” (PBDs), with clinical features being most severe in ZS and least pronounced in IRD. Inheritance of these disorders is autosomal recessive. The peroxisome biogenesis disorders are genetically heterogeneous, having at least 12 different complementation groups (CGs). The gene affected in CG1 is PEX1. Approximately 65% of the patients with PBD harbor mutations in PEX1. In the present study, we used SSCP analysis to evaluate a series of patients belonging to CG1 for mutations in PEX1 and studied phenotype-genotype correlations. A complete lack of PEX1 protein was found to be associated with severe ZS; however, residual amounts of PEX1 protein were found in patients with the milder phenotypes, NALD and IRD. The majority of these latter patients carried at least one copy of the common G843D allele. When patient fibroblasts harboring this allele were grown at 30°C, a two- to threefold increase in PEX1 protein levels was observed, associated with a recovery of peroxisomal function. This suggests that the G843D missense mutation results in a misfolded protein, which is more stable at lower temperatures. We conclude that the search for the factors and/or mechanisms that determine the stability of mutant PEX1 protein by high-throughput procedures will be a first step in the development of therapeutic strategies for patients with mild PBDs.
Introduction
Peroxisomes are subcellular organelles, present in virtually every eukaryotic cell, that catalyze a variety of essential functions. In humans, these functions include fatty-acid β-oxidation, etherphospholipid biosynthesis, fatty-acid α-oxidation, isoprenoid biosynthesis, and other functions (van den Bosch et al. 1992; Wanders and Tager 1998). Disorders resulting from defects in peroxisomal biogenesis include Zellweger syndrome (ZS [MIM 214100]), neonatal adrenoleukodystrophy (NALD [MIM 202370]), infantile Refsum disease (IRD [MIM 266510]), and rhizomelic chondrodysplasia punctata type 1 (RCDP1 [MIM 215100]) (Lazarow and Moser 1995; Wanders 1999). The first three diseases, listed in order of decreasing severity, represent a continuum of clinical features, whereas RCDP displays a distinct phenotype (Moser et al. 1995). Patients with ZS show neurological and hepatic abnormalities, renal cysts, and characteristic facial dysmorphic stigmata; these patients die at ages <1 year. Patients with IRD display the mildest phenotype, with no significant cerebral abnormalities, and survive until or even beyond adolescence (Lazarow and Moser 1995). NALD is intermediate between ZS and IRD in severity, and patients usually die in early childhood. In recent years, an increasing number of proteins, named “peroxins,” that are involved in the biogenesis of peroxisomes have been identified (Sacksteder and Gould 2000). Complementation studies with mammalian fibroblasts have so far shown the existence of 12 different groups; in 11 of these, the defective PEX gene is known (Moser 1999). Complementation group 1 (CG1) is the largest, characterized by mutations in HsPEX1 (PEX1 [MIM 602136]), a gene coding for a 143-kD AAA (ATPases associated with diverse cellular activities) protein (Portsteffen et al. 1997; Reuber et al. 1997; Tamura et al. 1998a). Approximately 65% of patients affected with peroxisome biogenesis disorders (PBDs [MIM 601539]) belong to this group (Moser et al. 1995; Maxwell et al. 1999; Wanders et al. 1999). The clinical phenotypes of patients belonging to this group cover a spectrum of severity, ranging from ZS to NALD and IRD.
With respect to the molecular basis of these phenotypes, two relatively common alleles in the HsPEX1 gene have been identified: one allele with a missense mutation (G843D) associated with milder PBDs (Portsteffen et al. 1997; Reuber et al. 1997; Imamura et al. 1998a; Collins and Gould 1999; Gärtner et al. 1999; Maxwell et al. 1999), and a second allele with a 1-bp insertion (c.2097-2098insT) resulting in a premature stop at amino acid 740, a mutation that is often present in patients with classical ZS (Collins and Gould 1999; Maxwell et al. 1999). Fibroblasts harboring the G843D allele are characterized by a temperature-dependent phenotype, with restoration of catalase import and peroxisomal functions at 30°C (Imamura et al. 1998a).
In the present study, we aimed to define the relationship between the genetic defects and the phenotypes of CG1 patients. For this purpose, genomic DNA from 22 Dutch patients belonging to CG1 was investigated by SSCP analysis and DNA sequencing. Subsequently, the amount of PEX1 protein in fibroblasts from these patients was determined. In addition, biochemical and cell-biological studies were performed to analyze the function of the PEX1 protein. Finally, we examined the way in which the G843D allele led to the observed temperature-dependent phenotype.
Patients, Material, and Methods
Patients
Clinical information about some of the patients included in the present study has already been reported in the literature. Patient 2 of our study is referred to as “patient 9” in the report by Heymans et al. (1984), patient 22 of our study is described in an article by Smeitink et al. (1992), and patients 27 and 30 of our study are referred to as “case 4” and “case 1,” respectively, in the report by Budden et al. (1986).
Cell Lines and Culturing Conditions
Primary fibroblast cell lines, from 22 Dutch patients who were assigned to CG1 by complementation analysis (Wanders et al. 1999) performed at the Amsterdam Medical Center, were cultured, at 8.5% CO2, in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (v/v) fetal calf serum, 2 mM l-glutamine, 100,000 U penicillin/liter, and 100 mg streptomycin/liter. Some of these cell lines were transformed with the large T antigen of SV40 virus, as described by Dodt et al. (1995).
To investigate the PEX1 protein level and the peroxisomal function under serum starvation, fibroblasts were seeded, in 75-cm2 flasks and on cover slips, to 50% cell confluency and were cultured in serum-free Opti-MEM (Life Technologies) for 3 d, before further analysis. In a separate experiment, subconfluent cultures, in flasks and on cover slips, were incubated with the proteasome inhibitor MG-132 (Calbiochem; final concentration 7.5 μM MG-132 and 0.03% [v/v] DMSO)—or with DMSO alone in DMEM plus supplements—for 6 h and then were processed. To evaluate the cells under highly confluent conditions, the cultures in flasks and on cover slips were kept for 7 d after they reached confluency and then were used for analysis.
Immunofluorescence Microscopy
Fibroblasts were seeded on cover slips 1 d before the actual experiment. The cells were processed for indirect immunofluorescence, as described by Slawecki et al. (1995). Polyclonal sheep anti–human catalase antibodies were obtained from The Binding Site; labeled secondary antibodies were acquired from standard commercial sources. Immunofluorescence was observed with a Zeiss Axiophot microscope.
Preparation of Cell Pellets, Western Blotting, Generation of Antibodies, and Biochemical Assays
Cells from a 75-cm2 flask, grown to near confluency, were harvested with trypsin, washed twice with Hanks buffer, and resuspended in 150 μl H2O. An aliquot was taken for protein estimation (Coomassie Protein Assay Reagent, Pierce). The cell suspension was immediately mixed with 5× concentrated SDS sample buffer (final concentration 2% (w/v) SDS, 10% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, 0.002% (w/v) bromophenol blue, and 62.5 mM Tris/HCl, pH 6.8), boiled for 5 min, and frozen at −80°C.
The cell lysates were separated by SDS-PAGE on 8% gels (25 μg total protein/slot). The separated proteins were transferred onto nitrocellulose membrane for 5 h at 30 V, using a BioRad mini blot apparatus (BioRad). Western blot analysis was performed with anti-PEX1 polyclonal rabbit antibodies (1:15,000 dilution) and with anti–rabbit IgG antibodies conjugated to horseradish peroxidase as secondary antibodies (Amersham), using the ECL western blotting detection reagent (Amersham). The exposed film (Hyperfilm ECL, Amersham) was digitized with a flatbed scanner (Arcus II, Agfa), and PEX1 protein was quantified by MacBas software (Fuji).
Anti-PEX1 antibodies were generated against the N-terminal part of PEX1, corresponding to amino acids 2–315. For this procedure, a 945-bp fragment was amplified by PCR, using the primer pairs KU 397 (5′-TCGGGCCCATGGGATGGGGCAGCGATCGCCTG-3′) and KU 398 (5′- CTGAGCTCGGATCCAAATACATGAATGGCACAG-3′), digested with NcoI and BamHI, and cloned downstream of the hexahistidinyl (His6) tag into the corresponding sites of expression plasmid pET9dNHis6. The latter plasmid was kindly provided by G. Stier (EMBL, Heidelberg). It was derived from pET9d (Novagen) by replacing the unique NcoI site with a DNA fragment that encoded the His6 tag and contained several unique endonuclease recognition sites, including NcoI. Tagged PEX1 proteins expressed in Escherichia coli BL21 (DE3) were purified by Ni-NTA chromatography under denaturing conditions (8 M urea), according to the pET system manual.
The quantification of very-long-chain fatty acids (VLCFA) and the determination of dihydroxyacetone-phosphate acyltransferase (DHAPAT), catalyzing the first step in etherphospholipid biosynthesis, were performed as described by Wanders et al. (1995) and Vreken et al. (1998).
Mutation Analysis
Genomic DNA was prepared as described by Miller et al. (1988), using 1 ml of lysis buffer (10 mM Tris/HCl, pH 8.2; 400 mM NaCl; and 2 mM EDTA) for a confluent 75-cm2 flask of fibroblasts. PCR was performed with intronic primer pairs encompassing exons 1–24 (see table 1). SSCP analysis was performed as described by Portsteffen et al. (1997). Alternatively, the SSCP reactions were performed without radioactive labeling, and the samples were separated on a 0.5 × MDE gel (FMC), containing 2% glycerol, 1 M urea, and 0.6 × TBE on GelBond PAG film (FMC). The DNA was visualized by silver staining. PCR fragments showing a mobility shift were directly sequenced. To this end, the PCR fragments were separated on agarose and isolated. Direct sequencing was performed by Qiagen.
Table 1.
Intronic Primer Pairs Used to Amplify the HsPEX1 Exons for SSCP Analysis[Note]
| Sequence | Designation | Sense (s) orAntisense (as) |
| 5′-CTTTGCGGCGCTAGGGT-3′ | 5inex1 | s |
| 5′-GGCTGAAGATCAGGTGGCTC-3′ | 3inex1 | as |
| 5′-AAAATGTATACGAAAACCTTTTTTCC-3′ | 5inex2 | s |
| 5′-ATGTGATATTAGAAGAAAGTTATTGC-3′ | 3inex2 | as |
| 5′-GATTACATAGACTACATATACACCC-3′ | 5inex3 | s |
| 5′-CATTGATATTGTGAAGTAATTTAACC-3′ | 3inex3 | as |
| 5′-ATTTTTTTTAACTAGATATGGAGTGG-3′ | 5inex4 | s |
| 5′-GCACATTCATTTCTACTTTGGACTC-3′ | 3inex4 | as |
| 5′-AGAAATGAATGTGCACTAATGAGC-3′ | 5inex5 | s |
| 5′-TAAGAATTCGGAAATACATGAATGGCACAG-3′ | 3exex5 | as |
| 5′-CGCGTCAGCAACCTCTG-3′ | 5exex5 | s |
| 5′-TGTTTAAGCCACATAAAATTTCTCC-3′ | 3inex5 | as |
| 5′-TAGAAGCTAATATATCAATATTTATGC-3′ | 5inex6 | s |
| 5′-CAACTAGAAATCTTACAAAACGTG-3′ | 3inex6 | as |
| 5′-GTTAAAGTTAAGTTCAGATATGAGG-3′ | 5inex7 | s |
| 5′-TACAACTATTTAAATAGAAAAAAAAGTC-3′ | 3inex7 | as |
| 5′-CTCTGTTTCAGTACTAACTCTGC-3′ | 5inex8 | s |
| 5′-CAAGGTGTTACAAGGAACTTCATA-3′ | 3inex8 | as |
| 5′-AAGACATACTTTGAAATGTAAAACTG-3′ | 5inex9 | s |
| 5′-TTGACATTAACATCTACTTTAATATTTAC-3′ | 3inex9 | as |
| 5′-TCAGTCTTTTATCATGTAACTATGTAT-3′ | 5inex10 | s |
| 5′-TATTAAATGTTACAGAAAAATGAACAC-3′ | 3inex10 | as |
| 5′-CTGAATCTTGGTGGTTGCC-3′ | 5inex11 | s |
| 5′-GATATGTGTATTTATTAGATTGACAG-3′ | 3inex11 | as |
| 5′-GCACTGAAATGATACTGAAACCAT-3′ | 5inex12 | s |
| 5′-GACTGACAAAAGAACAAGACCTTA-3′ | 3inex12 | as |
| 5′-TACTTTTCCTAAGCTTTTGCACTA-3′ | 5inex13 | s |
| 5′-TATAAAAGGGACATAATTCAATAATC-3′ | 3inex13 | as |
| 5′-CTGATTTTCTCCAAATATACATTCAA-3′ | 5ines14 | s |
| 5′-TAGAAAGAAGATTCCAAGTTCAGG-3′ | 3inex14 | as |
| 5′-TATAGTGAATAATAACTAGTAAAAGAAG-3′ | 5inex15 | s |
| 5′-CATAAAGCCAAAGCCAATAATACAG-3′ | 3inex15 | as |
| 5′-TTAGTCATTTCTGTTTAGACTTGAG-3′ | 5inex16 | s |
| 5′-TTTACACTTTGAAATGGCTAACTG-3′ | 3inex16 | as |
| 5′-ATTTCCATGATTCATTTACACTTAG-3′ | 5inex17 | s |
| 5′-TGATTGATAAATAATAACAGAGTCAG-3′ | 3inex17 | as |
| 5′-ACTTTGCCAACTATGAAGCCTG-3′ | 5inex18 | s |
| 5′-ACCAAAATCTGATGACATGATGAC-3′ | 3inex18 | as |
| 5′-GTCATCATGTCATCAGATTTTGGT-3′ | 5inex19 | s |
| 5′-TAGCATTTGTTGGGTTTTGGAC-3′ | 3inex19 | as |
| 5′-ATTTATGAAGAAAATCATAAGATACAC-3′ | 5inex20 | s |
| 5′-TGACATTGTACTTCTTTTATCACTC-3′ | 3inex20 | as |
| 5′-TAAAATTGTTTACATCTTTAAACTGG-3′ | 5inex21 | s |
| 5′-AGAAATCACTGCAACTTTACACC-3′ | 3inex21 | as |
| 5′-GTTTTTTCTCTCCCCCTCTTCC-3′ | 5inex22 | s |
| 5′-TAAGAAGTTTTAACAATTATAATGAGG-3′ | 3inex22 | as |
| 5′-TTTTACATCCTTAATTTAACTCTTCG-3′ | 5inex23 | s |
| 5′-ACTTGTAATAGTAGCTGTACTTCC-3′ | 3inex23 | as |
| 5′-ATCCATTATCTTTTGTTTTGTAATGG-3′ | 5inex24 | s |
| 5′-CTGTTACAACATATGGAAAAGCC-3′ | 3inex24 | as |
Note.— Examples for designations: 5inexE is the sense primer attaching to the 5′ end of exon E, and 3inexE is the antisense primer attaching to the 3′ end of exon E.
Results
PEX1 Mutations in Patients from CG1 with Different Clinical Phenotypes
One goal of this study was to investigate the genetic differentiation of severely and mildly affected patients. Although the biochemical diagnosis of the PBDs is well established, the correlation between the biochemical data and the clinical presentation is still unclear. We analyzed 22 patients, belonging to CG1, who had different clinical phenotypes for mutations in PEX1. Each of the 24 PEX1 exons were amplified from fibroblast DNA through use of intronic primer pairs and analyzed by SSCP (Portsteffen et al. 1997). Fragments showing unusual patterns after electrophoretic separation were sequenced directly, in both directions. All mutations are depicted in table 2.
Table 2.
Relationships between Genotype, Clinical Phenotype, Biochemical Parameters, and the PEX1 Protein Level, in Fibroblasts from Patients of CG1
|
Mutation (cDNA Level, Protein Level) |
||||||
| Disease Categoryand Patient | I | II | Age at Deathor at Present | PEX1 Protein Level(% of control) | VLCFAC26:C22 Ratioa | DHAPAT Activityb(nmol/2h × mg protein) |
| Nonclassical ZS: | ||||||
| 2 | c.2528G→A, G843D | c.2528G→A, G843D | 2 years, 9 mo | ∼3 | .45 | .5 |
| 22 | c.2528G→A, G843D | c.2528G→A, G843D | 9 years | <10 | .30 | 2.1 |
| 26c | c.2528G→A, G843D | c.2528G→A, G843D | 8 years, alive | ∼15 | .18 | .2 |
| 27c | c.2528G→A, G843D | c.2528G→A, G843D | 27 years, alive | ∼10 | .12 | 2.4 |
| 31c | c.2528G→A, G843D | c.2528G→A, G843D | 45 years, alive | ∼7 | .49 | 1.4 |
| 10c | c.2528G→A, G843D | c.2097-2098insT, P740X | 41 years, alive | ∼5 | .29 | 1.2 |
| 13c | c.2528G→A, G843D | c.2097-2098insT, P740X | 5.5 years, alive | <5 | .39 | .6 |
| 20 | c.2528G→A, G843D | c.2097-2098insT, P740X | 9 years | 5–10 | .27 | 1.9 |
| 28 | c.2528G→A, G843D | c.2097-2098insT, P740X | 2.5 years | 5–10 | .70 | 1.5 |
| 30c | c.2528G→A, G843D | c.2097-2098insT, P740X | 21 years, alive | <5 | .18 | 1.4 |
| 7d | c.2528G→A, G843D | 3850T→C, X1284Q | Unknown, >2 years | ∼3 | .28 | nde |
| 9c | c.1777G→A, G593R | c.2071+1G→T | 6 years, alive | ∼7 | .12 | 4.3 |
| Mean ± SD | .31±.16 | 1.59±1.07 | ||||
| Classical ZS: | ||||||
| 1 | c.2528G→A, G843D | c.2846G→A, R949Q | 3 mo | ∼50 | .34 | 2.0 |
| 17 | c.2097-2098insT, P740X | c.2097-2098insT, P740X | 2 mo | 0 | .52 | .5 |
| 19 | c.2097-2098insT, P740X | c.2097-2098insT, P740X | Unknown | 0 | .32 | 1.1 |
| 23 | c.2097-2098insT, P740X | c.2097-2098insT, P740X | 1 year | 0 | .60 | .5 |
| 6 | c.788-789delCA, T268X | c.2097-2098insT, P740X | 3 mo | 0 | .73 | .5 |
| 16 | c.2926+2T→C | c.2097-2098insT, P740X | Unknown | 0 | .33 | .5 |
| 11 | c.2730delA, V960X | c.2730delA, V960X | 3 mo | 0 | .31 | .1 |
| 12 | c.2730delA, V960X | c.2730delA, V960X | 3 mo | 0 | .48 | .1 |
| 14 | c.1670+5G→T | c.1670+5G-T | 18 mo | 0 | .61 | .1 |
| 5 | c.434-448delf | c.2008C→A, L670M | 1 day | 0 | .59 | .4 |
| Mean ± SD | .48±.14 | .58±.55 | ||||
Control values: mean = .04; range = .03–.07.
Control value: 8.5 ± 2.0 nmol/2h × mg protein.
Patient's phenotype is IRD.
Patient's phenotype is NALD.
nd = not determined.
Patient has c.434-448del and insertion of GCAA.
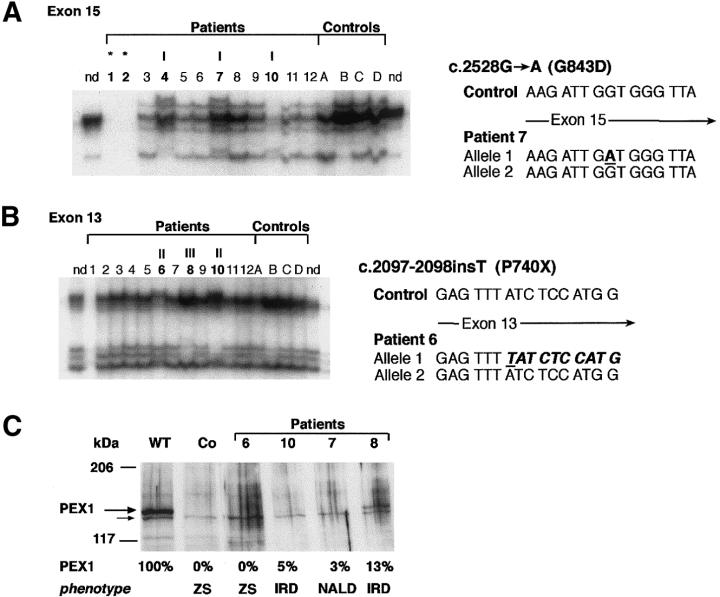
Approximately half of the alleles (21 of 44) carry a missense mutation, with a clear prevalence of the G843D allele (17 of 44 alleles; table 2). Apart from this allele, we identified one other frequent allele, the c.2097-2098insT allele, which harbors an insertion of a nucleotide in exon 13 leading to a premature stop at codon 740 (P740X). We found the c.2097-2098insT mutation in 13 of 44 alleles; this means that the c.2097-2098insT allele has a frequency of 30% among the Dutch patients with PBD who belong to CG1. Since we observed that this allele was not always detected by SSCP analysis, we reinvestigated the six patients included in our previous study (Portsteffen et al. 1997) by means of direct sequencing and found five of the patients to be heterozygous for this c.2097-2098insT allele (Reuber et al. 1997; Collins and Gould 1999). The c.2730delA allele, which results in a premature stop at codon 960 (V960X), was identified in two related patients who were both homozygous for the 1-bp deletion. The SSCP analysis and an example of the sequencing results are given in figure 1A, for exon 15, and in figure 1B, for exon 13. The c.2730delA allele is described in figure 2B and in table 2.
Figure 1.
Common mutations of CG1, and resulting PEX1 protein levels. A, The G843D allele (I), identified in patients 1, 2, 4, 7, and 10 by SSCP analysis and sequencing of exon 15. The genotypes of patients indicated by an asterisk (*) were identified by sequence analysis. nd = not denatured control. Numbers 1–12 indicate CG1 patients 1–12. B, The c.2097-2098insT allele (indicated by “II”), detected in patients 6 and 10 by SSCP analysis and sequencing of exon 13. The frameshift in one allele leads to a premature stop at codon 740 (P740X). “III” indicates a newly identified HsPEX1 allele in patient 8, which results in I696M. C, Results of immunoblot analysis using polyclonal antibodies against PEX1 protein. Patients 7 and 10, carrying the G843D allele, had residual PEX1 protein (∼5% of the normal level) and were affected with the milder phenotypes NALD or IRD (i.e., nonclassical ZS). No PEX1 was detected in cell lysates from patient 6, who was carrying the c.2097-2098insT allele; this patient had severe classical ZS. The PEX1 protein level in fibroblasts from patient 8 is shown for comparison. WT = wild type; Co = control fibroblasts from patient PBD009, who has no detectable PEX1 mRNA and who exhibits ZS (Collins and Gould 1999); smaller arrow indicates unspecific protein recognized by the anti-PEX1 antibodies.
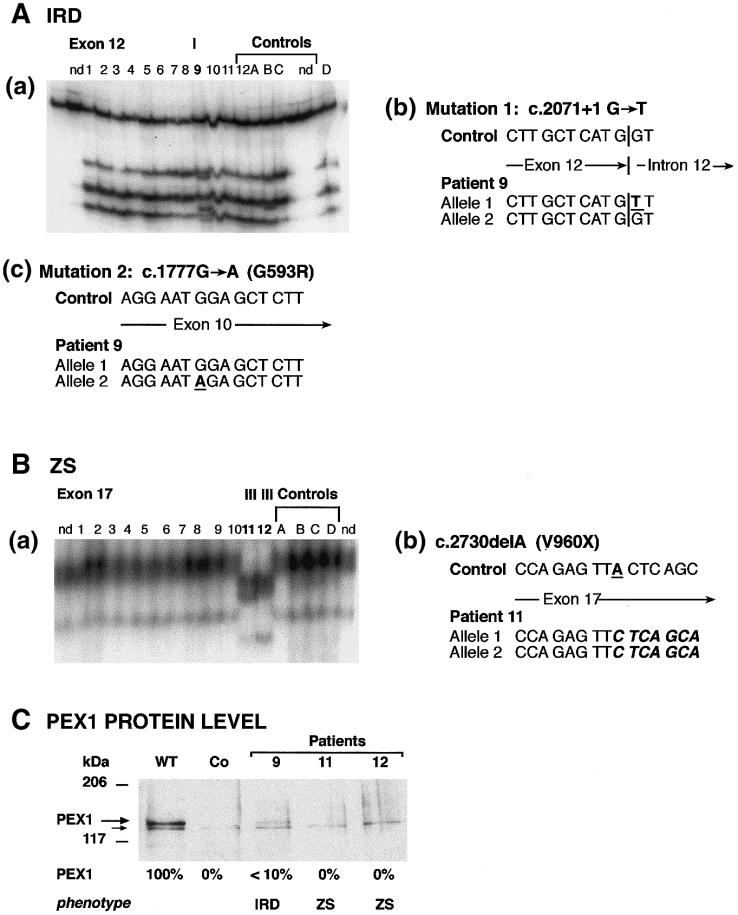
Figure 2.
Genotypes and PEX1 protein levels in patients with IRD and ZS. Abbreviations are as in figure 1. A, Patient 9, affected with IRD: (a) SSCP analysis of exon 12 showed a mobility shift (indicated by “I”); nd = not denatured control, numbers 1–12 indicate CG1 patients 1–12; (b) sequencing of exon 12 led to identification of a G→T transversion at the donor splice site (c.2071+1G→T) in one allele; (c) sequencing of exon 10 revealed one c.1777G→A allele, resulting in G593R. B, Patients 11 and 12, affected with classical ZS: (a) SSCP analysis of exon 17 showed a mobility shift; (b) sequencing revealed a 1-bp deletion of nucleotide 2730 (c.2730delA), resulting in a stop codon at amino acid 960 (V960X). C, The PEX1 protein levels, investigated in cell lysates of patient fibroblasts, given as percentages of the wild-type level. No PEX1 was found in cells of the patients with classical ZS (patients 11 and 12). Approximately 10% of the normal level of PEX1 protein was detected in cells from patient 9, who was affected with the mild clinical phenotype IRD (nonclassical ZS).
Correlation between Different Mutant Alleles and the Resulting PEX1 Protein Levels
On the basis of our assumption that the phenotypes of the patients correlate with the degree of peroxisomal function and thus depend on functional PEX1 protein, we investigated the steady-state levels of PEX1 protein in fibroblasts. For this purpose, we successfully generated antibodies against PEX1. These antibodies were directed against amino acids 2–317 of HsPEX1 and therefore also recognize truncated proteins.
Fibroblasts from all patients were harvested and lysed in sample buffer. Proteins were separated by SDS-PAGE, blotted onto nitrocellulose, and probed with polyclonal anti-PEX1 antibodies. The consequences of the two most common alleles for the PEX1 protein level are shown in figure 1C and table 2.
Instead of the typical categories of ZS, NALD, and IRD, we decided to use only two disease categories, comprising (1) classical ZS (craniofacial dysmorphia; severe neurological abnormalities, including hypotonia; eye abnormalities; and early death) and (2) nonclassical ZS, which includes all variants that are not classical ZS. This second group mainly represents patients with a diagnosis of NALD and IRD, who show less-pronounced abnormalities and have longer survival. The designation “NALD” and “IRD” was also given, for those patients in whom the diagnosis was well defined.
All fibroblasts from patients harboring the G843D allele had amounts of residual PEX1 protein that were 3%–15% of that found in normal fibroblasts (table 2A). The clinical presentations of these patients indicated milder phenotypes (i.e., nonclassical ZS) like NALD or IRD, and survival times were >2 years. An exception was patient 1, who exhibited ∼50% PEX1 immunoreactive protein but nevertheless showed all the signs and symptoms of classical ZS. It is possible that the second allele, R949Q, is expressed in this patient but is nonfunctional, and that it interferes with the residual protein expressed from the G843D allele.
No protein was expressed from the c.2097-2098insT (P740X) allele, as demonstrated by analysis of patients 17, 19, 23, 6, and 16 (table 2B). These findings are in line with the results of Maxwell et al. (1999), who detected almost no PEX1 mRNA. All patients who were either homozygous for this allele—or heterozygous for this allele, with the other allele being a different “null allele”—had no detectable PEX1 protein and were all diagnosed with classical ZS. For two patients, the age at death is unknown because there was no further follow up, owing to different reasons. All other patients died within the first 2 years of life.
Two additional examples of patients with a combination of different mutant alleles also demonstrate the effect of the mutations on the PEX1 protein level (fig. 2). Patient 9 was heterozygous for an allele with a splice-donor–site mutation in exon 12 (c.2071+1G→T) and for an allele with a missense mutation, resulting in G593R (fig. 2A). As seen in figure 2C, the protein level was <10% of that in wild-type fibroblasts. The allele carrying the splice-site mutation is likely to produce unstable RNA and, probably, no protein (Maquat 1995). No abnormally sized protein band was visible with anti-PEX1 antibodies. Most likely, the G593R allele accounts for the residual PEX1 protein detected (∼7% of normal) in this patient with nonclassical ZS (IRD). Two identical twins (patients 11 and 12) were homozygous for the deletion of nucleotide 2730 (c.2730delA; fig. 2B). No PEX1 protein was detectable in fibroblasts from these patients. As mentioned above, this deletion would lead to a premature stop at amino acid 960. No abnormally sized protein bands reacting with the PEX1 antibodies were observed.
Since the presence of detectable PEX1 protein does not guarantee its functionality, as shown above for patient 1, we included biochemical parameters. Although the function of PEX1 is not defined yet, it is likely that PEX1 plays a significant role in the import of peroxisomal matrix proteins into the peroxisomes (Portsteffen et al. 1997; Reuber et al. 1997; Collins et al. 2000; Sacksteder and Gould 2000). As a direct measure of PEX1 activity in cells, we determined (1) the activity of the peroxisomal enzyme DHAPAT, and (2) the VLCFA levels (ratio of C26:C22) in fibroblasts. These are indicators of two distinct biochemical pathways: plasmalogen biosynthesis and peroxisomal β-oxidation. The results of biochemical analyses for all patients are summarized in table 2.
As can be judged from the mean values given in table 2, there is a clear tendency for a more severe biochemical phenotype for the patients with classical ZS. However, it is not reliable to assign a patient to one of the groups (nonclassical ZS or classical ZS) solely on the basis of these two biochemical parameters. In this regard, the additional determination of the protein level in combination with the mutation analysis will be more valuable. An example of the combination of these data is given in table 3, for patient 9 versus patients 11 and 12. Judging from VLCFA levels, DHAPAT activity, and partial localization of catalase in peroxisomes, it can be concluded that patient 9 seems to be mildly affected. These observations are in line with the diagnosis of nonclassical ZS (IRD). Patients 11 and 12, who lacked PEX1 completely, exhibited fully developed classical ZS with no import of peroxisomal matrix proteins, strongly elevated levels of VLCFA, almost no DHAPAT activity, and death before age 3 mo. The effect of a total deficiency of PEX1 protein was similar in all patients (see table 2) and always resulted in classical ZS.
Table 3.
Biochemical Parameters Correlated with the PEX1 Protein Level
| Biochemical Parameter | Patient 9 | Patient 11/Patient 12 |
| VLCFA C26:C22 ratioa | .12 | .31/.48 |
| DHAPAT activityb (nmol/2h × mg protein) | 4.3 | .1/.1 |
| Catalase import | Partial import into peroxisomes | No import into peroxisomes |
| Age/condition | 6 years, alive, relatively good condition | Died before age 3 mo |
| PEX1 protein level (% of control) | ∼7 | 0/0 |
| Diagnosis | Nonclassical ZS (IRD) | Classical ZS |
Control values: mean = .04; range = .03–.07.
Control values: 8.5 ± 2.0 nmol/2h × mg protein.
Elevation of the PEX1 Protein Level in Fibroblasts Harboring the G843D Allele, at 30°C
When we examine the PEX1 protein levels in all patient fibroblasts with the G843D allele, the steady-state level is reduced to 5%–10% of that in control fibroblasts. Since the mRNA level in these patients seemed to be normal (Maxwell et al. 1999), a likely possibility is that the mutant PEX1 protein is degraded immediately. It has been reported that missense mutations often result in partially misfolded proteins that undergo rapid degradation (Waters et al. 1998a, 2000; Bross et al. 1999). The possibility that the PEX1 G843D protein is less stable is supported by the temperature-sensitive phenotype that has been described for fibroblasts harboring this allele (Imamura et al. 1998a). Such a temperature effect is a rather common feature of proteins that arise from alleles with missense mutations, since misfolding is reduced at the lower temperature (Bross et al. 1999). This results in higher protein levels, leading to an almost normal phenotype. To test this prediction, we investigated the PEX1 function by monitoring the import of catalase and by analyzing the PEX1 protein level under various conditions. The following conditions were chosen to test the stability: (1) cells were cultured at a temperature of 30°C and were compared to cells cultured at 37°C; (2) the growth rate of the culture was modified by serum starvation; (3) the confluency of the fibroblasts was varied; and (4) cells were grown in the presence of proteasome inhibitors. Conditions 2 and 3 were selected because it has been reported that differences in peroxisomal import of mutant cell lines depend on the growth rate (serum starvation) or the confluency of the culture (Wiemer et al. 1991).
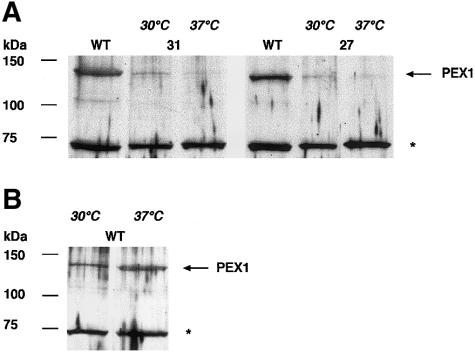
We selected cells from patients 10, 26, 27, and 31, who were all homozygous or heterozygous for the G843D allele, together with control cells (table 4). We first investigated the temperature effect on the import of catalase. Catalase is a sensitive indicator of peroxisomal-import defects because of its very inefficient import even under normal conditions (Lazarow et al. 1982; Warren et al. 1998). Specifically, we cultured fibroblasts from patients and a control for 3 d at 37°C and at 30°C. The overall results are listed in table 4. Examples of the immunoblots with PEX1 proteins are given in figure 3, and the immunofluorescence results are shown in figure 4.
Table 4.
Higher PEX1 Protein Levels, in Cells Harboring the G843D Allele, at 30°C Compared with 37°C
|
Mutation (cDNA Level, Protein Level) |
PEX Protein Level(% of control) |
Catalase Importa |
|||||
| Patient | I | II | At 37°C | At 30°C | Diagnosis and Condition | At 37°C | At 30°C |
| 10 | c.2528G→A, G843D | c.2097-2098insT, P740X | ∼5 | ∼17 | Nonclassical ZS (IRD), aged 41years, alive | (+) | +++ |
| 26 | c.2528G→A, G843D | c.2528G→A, G843D | ∼15 | ∼28 | Nonclassical ZS (IRD), aged 8 years, alive | + | ++++ |
| 27 | c.2528G→A, G843D | c.2528G→A, G843D | ∼10 | ∼18 | Nonclassical ZS (IRD), aged 27 years, alive | + | ++++ |
| 31 | c.2528G→A, G843D | c.2528G→A, G843D | ∼7 | ∼24 | Nonclassical ZS (IRD), aged 45 years, alive | + | ++++ |
| 1 | c.2528G→A, G843D | c.2846G→A, R949Q | ∼50 | ∼60 | Classical ZS; died at age 3 mo | … | (+) |
| Control | … | … | 100 | 80–90 | … | ++++ | ++++ |
… = no catalase import, (+) = 10% catalase import, + = 25% catalase import, +++ = 75% catalase import, and ++++ = 100% catalase import.
Figure 3.
Increase in the PEX1 protein level of patient fibroblasts harboring the G843D allele, at 30°C. Cells were cultured for 3 d at 37°C or 30°C. Cell lysates were subjected to SDS-PAGE and transferred onto nitrocellulose. Immunoblot analysis was performed, using polyclonal antibodies against PEX1 protein. WT = wild type (GM5659); “31” and “27” refer to patients 31 and 27, respectively. An arrow (←) marks the position of the PEX1 protein; the asterisk (*) denotes an unspecific protein recognized by anti-PEX1 antibodies, which was used to correct for an eventually unequal loading of the sample. The protein levels are shown in table 4. A, Comparison of wild-type (GM5659) cells versus cells from patients 31 and 27, at 30°C and 37°C. In comparison with wild-type cells, we detected ∼5%–17% of the normal level of PEX1 protein in lysates of cells incubated at 37°C, and ∼17%–28% of the normal level of PEX1 protein in those incubated at 30°C (see table 4 for protein levels). This increase in protein levels may be explained by an improved stability of PEX1 at the lower temperature. B, Lower PEX1 protein level in control wild-type fibroblasts at 30°C compared with that at 37°C, under the same conditions as above.
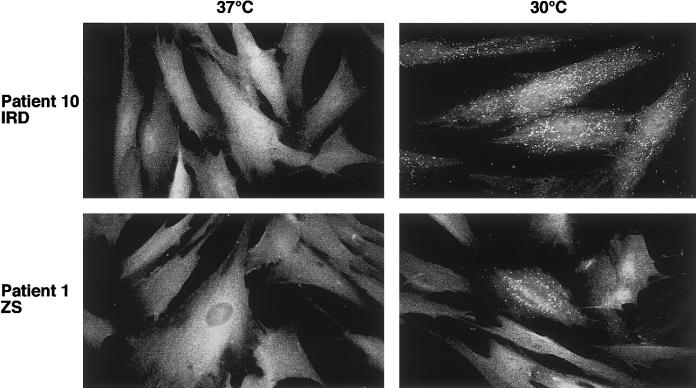
Figure 4.
Catalase import in peroxisomes of patient fibroblasts harboring the G843D allele and different second mutated PEX1 alleles. Fibroblasts were cultured for 3 d at either 30°C or 37°C and then treated for indirect immunofluorescence. Catalase staining was diffuse, indicating a cytoplasmic localization at 37°C. At 30°C, catalase was imported into peroxisomes in most of the cells from patient 10 (as indicated by the punctuate staining pattern) but in only ∼10% of the cells of patient 1. The punctuate structures colocalized with the staining for the peroxisomal membrane protein PMP70 (data not shown).
In fibroblasts from patients 10, 26, 27, and 31, the PEX1 protein level was two- to threefold higher at the lower temperature than at the higher temperature, in contrast to control cells, which even showed slightly reduced PEX1 protein levels at the lower temperature (table 4). Each cell line was investigated at least twice, with similar results. At this temperature, the import capabilities for catalase were completely restored. All these patients share a milder clinical phenotype and relatively extended survival times; patient 1, however, displayed a different picture. Although the PEX1 level in the cells from this patient, at the lower temperature, rose from 50% (at 37°C) to 60% of the normal level, the effect on the restoration of catalase import was only minimal. The clinical phenotype of this patient is described as classical ZS, with death at age 3 mo. As mentioned above, the second allele (R949Q) seems to be expressed but the protein may be nonfunctional and may interfere with the mutant protein from the G843D allele, thus preventing the alleviating effect of this allele.
In the second experiment, we investigated whether the observed effect of temperature (the higher import rate of catalase) may be related to the lower growth rate at 30°C, as has been reported for certain cell lines (Wiemer et al. 1991). We repeated the experiment described above, for patient 31 and for normal fibroblasts, with serum-free medium. This led to a substantial reduction in the growth of the investigated cells. Whereas the import in cells that were grown at 30°C in serum-free medium was restored, no restoration was observed in cells that were grown at 37°C in serum-free medium. Normal fibroblasts showed no import abnormalities under these conditions.
In the third experiment we compared cells—from patients 27 and 31 and from controls—that were at initial confluency versus cells that had been left completely confluent for 1 wk, and we then investigated catalase import and the PEX1 protein level. We observed a slight increase in catalase import and a minimal increase in the PEX1 level in all patient cells; however, the effect was far less pronounced than that observed in the first experiment characterized by the temperature change (data not shown).
Slower degradation of protein would result from a lower activity of proteases or of the protein-degrading machinery. To test this possibility, we incubated normal fibroblasts and fibroblasts from patient 26 for 6 h in 7.5 μM MG-132, a proteasome inhibitor, and investigated catalase import by immunofluorescence and the PEX1 protein level by immunoblotting. For control purposes, we analyzed the PEX5 (MIM 600414) protein level, which is severely reduced in PEX1-deficient cells, owing to higher degradation, as demonstrated by pulse-chase experiments (Dodt and Gould 1996). We noticed an increase in PEX5 protein level in the presence of the proteasome inhibitor, indicating that the concentration of MG-132 was, in principle, sufficient to block the degradation of PEX5. However, we could not detect any significant increase in PEX1 protein level. In addition, there was no apparent improvement in catalase import. This experiment was repeated with different PEX1 deficient cell lines, with similar results (data not shown).
Discussion
In principle, we can identify two groups of patients within CG1, as studied in this article. In the first group, there is a clear relationship between genotypes that are, presumably, “null alleles,” and the classical ZS phenotype, with death of the patients at ages <1–2 years. In these cases, we could not identify any PEX1 protein in fibroblasts, the import for peroxisomal proteins was almost completely absent, and the peroxisomal functions were severely reduced.
The second group includes all patients with genotypes that allow residual function of the PEX1 protein and who display severe yet milder phenotypes of nonclassical ZS, such as NALD and IRD. On the basis of our observations to date, the function of the peroxisome in single cells—for example, the import of catalase—depends on the amount of mutated PEX1 protein and on gene dosage. We observed the tendency that all fibroblasts of patients with at least one G843D allele have PEX1 protein levels that are ∼5% of that found in wild-type fibroblasts. Some of the patients with two G843D alleles have a higher amount of protein and are less severely affected. In spite of this correlation, it is not possible to make reliable predictions, regarding the prognosis of the milder affected patients, on the basis of the protein levels themselves or from other routinely determined biochemical data.
All mutant PEX1 alleles currently known, from the present study and from other published studies, are summarized in figure 5. The two common alleles G843D and c.2097-2098insT account for 39% and 30%, respectively, of the CG1 alleles in our patients. This roughly corresponds to the published results for patients at the Kennedy Krieger Institute in Baltimore (Collins and Gould 1999). These authors described an allele frequency of 30% each, for both mutations. In addition, frequencies of 25% for the G843D and 32% for the c.2097-2098insT have been reported for Australasian probands (Maxwell et al. 1999). Other published frequencies include 33% for the G843D allele in a German cohort (Gärtner et al. 1999) and 21% for this same allele in probands that were examined by a Japanese group (Imamura et al. 1998a).
Figure 5.
Mutations in PEX1. A, Exons coding for the first AAA cassette of PEX1, depicted in light gray, and exons coding for the second AAA cassette, in dark gray. Mutations identified by our group (28 CG1 patients; Portsteffen et al. 1997; and the present study) are marked above the row of exons, and mutations identified by other groups (Reuber et al. 1997; Imamura et al. 1998a; Tamura et al. 1998a; Collins and Gould 1999; Gärtner et al. 1999; Maxwell et al. 1999) are marked below the row of exons. Gray symbols = cell lines from identical patients were analyzed by different groups. Each symbol represents a single PEX1 allele. The numbers refer to the references given below. B, Descriptions of the mutations, sorted by exon, with designations given at the cDNA and the protein level whenever possible. A silent mutation at codon 777 (G777) that seems to be a polymorphism, as well as an intronic deletion in intron 11, both of which are described by Collins and Gould (1999), are not included. s.d. = splice donor site. References—1 = Reuber et al. (1997); 2 = Portsteffen et al. (1997); 3 = Gärtner et al. (1999); 4 = Tamura et al. (1998a); 5 = Maxwell et al. (1999); 6 = Collins and Gould (1999); 7 = Imamura et al. (1998a).
Misfolding as a Reason for Reduced PEX1 Protein Levels?
Recent data on genotype-phenotype relationships in phenylketonuria may be helpful in the interpretation of our results (Waters et al. 1998b, 2000). Waters and coworkers investigated several missense alleles by observing their expression in different in vitro systems and by 3D-structural localization. They found that some of the missense mutations that are changing amino acids far from the catalytic site of phenylalanine hydroxylase lead to misfolding of the protein monomers, to altered oligomerization into the tetramer, and to an accelerated proteolytic degradation, finally resulting in a reduced cellular protein level of the enzyme (Waters et al. 1998b, 2000). These mutant phenylalanine hydroxylases were associated with a variability of phenotypic severity that the authors explained by interindividual differences in the handling of the mutant proteins (Waters et al. 2000).
Our results for the G843D allele suggest that the amount of PEX1 protein and the peroxisomal functions in patient fibroblasts can be influenced by environmental conditions, such as incubation temperature or confluency of the culture. Furthermore, it is likely that, on a cellular level, ∼20% of the normal protein level is sufficient to allow normal catalase import. From this we cannot directly define the amount of PEX1 protein that would be necessary to prevent a clinical phenotype, but we do know that 5%–15% of normal levels of functional PEX1 protein is related to the milder clinical phenotypes. This indicates that very small changes in the protein level may be beneficial for patients.
In this respect, it is interesting to note that Singh et al. (1997) have demonstrated that the import of catalase is very important for the metabolic functions of peroxisomes. They suggest that more-mildly affected patients, especially, may import sufficient levels of other peroxisomal enzymes but have a more pronounced defect in catalase import. The function of the imported enzymes would be inhibited because of accumulation of hydrogen peroxide in catalase-negative peroxisomes.
The probability that the G843D PEX1 protein is unstable is in line with the following data: PEX6 (MIM 601498), the second AAA protein involved in peroxisome biogenesis, has been identified as an interacting partner for PEX1 by two-hybrid analysis and immunoprecipitation (Geisbrecht et al. 1998; Tamura et al. 1998b). In addition, the PEX phenotype of some PEX1-deficient fibroblasts can be partially overcome by expression of PEX6 in certain cell lines (Geisbrecht et al. 1998). Interestingly, the susceptible cell lines are the ones that carry the G843D mutation and thus have residual PEX1 protein. No suppression was observed in cell lines known to express no PEX1 protein. Similar results have been reported for the yeast Pichia pastoris (Faber et al. 1998). It seems likely that overexpression of the interacting protein PEX6 stabilizes some PEX1 protein, in a way similar to the effects of temperature shifting. The interaction between the G843D PEX1 protein (expressed at 37°C) and PEX6 is reduced by ∼70% compared with the interaction with the wild-type PEX1 and PEX6 (Geisbrecht et al. 1998). In addition, overexpression of the G843D allele in PEX1-deficient cells at 37°C suggested a reduced function (15% of normal activity) as compared with the wild-type allele (Reuber et al. 1997). If we postulate that the reduction in PEX1 abundance reflects an error of PEX1 folding and that anything that prevents this misfolding results in a greater amount of correctly folded and active PEX1, then the effects described above could be explained as follows: The overexpression of G843D PEX1 at 37°C leads to the synthesis of partially misfolded protein, which results in a reduced capability of interaction with PEX6 and reduced PEX1 function in general.
Misfolding as a Target for Pharmacological Treatment?
The difficulties encountered in an attempt to divide—by phenotype—patients who have nonclassical ZS and have the G843D allele into supgroups that have one or two copies of the G843D allele may be explained by the fact that the G843D PEX1 protein level depends on other factors (not only on gene dosage) involved in the folding and misfolding of the mutant protein. These factors include different levels of chaperones and differences in the proteolytic degradation of the mutant protein, both of which can be determined genetically or by environmental conditions. In addition, the temperature effect may be important and may represent a simple parameter to define further mutant alleles. The features of the G843D protein present the intriguing possibility that any actions that help to stabilize the residual protein may be used for some kind of therapy for patients with this kind of mutation. In this context, it is interesting to note that a recent study of the pharmacological induction of peroxisomes in PBDs described the different responses, to treatment with 4-phenylbutyrate, of cells from patients with ZS and cells from patients with NALD or IRD (Wei et al. 2000). Although improvement of peroxisomal function, measured by a two- to threefold increase in VLCFA oxidation and plasmalogen concentration, was observed in cells of patients with milder phenotypes, this was not the case in cells of patients with ZS. In addition, these authors showed that PEX1 and PEX6 mRNA was not increased in treated cells of patients with milder CG1 phenotypes. Since we assume that the improvement of peroxisomal function (VLCFA oxidation and plasmalogen synthesis) in these patient cells would require a gain in PEX1 function, 4-phenylbutyrate will be an interesting candidate for the investigation of pharmacological influence on PEX1 protein.
Almost 30% of all patients with PBD (excluding those with RCDP) carry at least one G843D allele. An additional missense mutation (G593R) associated with low PEX1 protein levels is a candidate for a similar phenotype. Imamura et al. (1998a, 1998b) have described additional mutations, in other complementation groups, with a cellular phenotype that is influenced by temperature changes. This means that a large group of mildly affected patients would probably benefit from therapeutic efforts to stabilize the protein. Therefore, pharmacological agents should be screened via high-throughput procedures. The observations of the present study provide a new target for causal pharmacological treatment of patients with milder PBD phenotypes.
Acknowledgments
We are grateful to S. Wüthrich and E. Becker for their assistance. G.D. received a Lise Meitner fellowship of the state North Rhine-Westphalia. This work was supported by Forschungsförderung Ruhr-Universität Bochum Medizinische Fakultät, by the Thyssen Stiftung, and by the Princess Beatrix Fonds, The Hague, The Netherlands.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
References
- Bross P, Corydon TJ, Andresen BS, Jorgensen MM, Bolund L, Gregersen N (1999) Protein misfolding and degradation in genetic diseases. Hum Mutat 14:186–198 [DOI] [PubMed] [Google Scholar]
- Budden SS, Kennaway NG, Buist NR, Poulos A, Weleber RG (1986) Dysmorphic syndrome with phytanic acid oxidase deficiency, abnormal very long chain fatty acids, and pipecolic acidemia: studies in four children. J Pediatr 108:33–39 [DOI] [PubMed] [Google Scholar]
- Collins CS, Gould SJ (1999) Identification of a common PEX1 mutation in Zellweger syndrome. Hum Mutat 14:45–53 [DOI] [PubMed] [Google Scholar]
- Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ (2000) The peroxisome biogenesis factors pex4p, pex22p, pex1p, and pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol 20:7516–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet 9:115–125 [DOI] [PubMed] [Google Scholar]
- Dodt G, Gould SJ (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S (1998) Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol 18:936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner J, Preuss N, Brosius U, Biermanns M (1999) Mutations in PEX1 in peroxisome biogenesis disorders: G843D and a mild clinical phenotype. J Inherit Metab Dis 22:311–313 [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Collins CS, Reuber BE, Gould SJ (1998) Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci USA 95:8630–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymans HS, van den Bosch H, Schutgens RB, Tegelaers WH, Walther JU, Muller-Hocker J, Borst P (1984) Deficiency of plasmalogens in the cerebro-hepato-renal (Zellweger) syndrome. Eur J Pediatr 142:10–15 [DOI] [PubMed] [Google Scholar]
- Imamura A, Tamura S, Shimozawa N, Suzuki Y, Zhang Z, Tsukamoto T, Orii T, Kondo N, Osumi T, Fujiki Y (1998a) Temperature-sensitive mutation in PEX1 moderates the phenotypes of peroxisome deficiency disorders. Hum Mol Genet 7:2089–2094 [DOI] [PubMed] [Google Scholar]
- Imamura A, Tsukamoto T, Shimozawa N, Suzuki Y, Zhang Z, Imanaka T, Fujiki Y, Orii T, Kondo N, Osumi T (1998b) Temperature-sensitive phenotypes of peroxisome-assembly processes represent the milder forms of human peroxisome-biogenesis disorders. Am J Hum Genet 62:1539–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow PB, Moser HW (1995) Disorders in peroxisome biogenesis. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited disease. McGraw Hill Inc., New York, pp 2287–2324 [Google Scholar]
- Lazarow PB, Robbi M, Fujiki Y, Wong L (1982) Biogenesis of peroxisomal proteins in vivo and in vitro. Ann NY Acad Sci 386:285–300 [DOI] [PubMed] [Google Scholar]
- Maquat LE (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1:453–465 [PMC free article] [PubMed] [Google Scholar]
- Maxwell MA, Nelson PV, Chin SJ, Paton BC, Carey WF, Crane DI (1999) A common PEX1 frameshift mutation in patients with disorders of peroxisome biogenesis correlates with the severe Zellweger syndrome phenotype. Hum Genet 105:38–44 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AB, Rasmussen M, Naidu S, Watkins PA, McGuinness M, Hajra AK, Chen G, Raymond G, Liu A, Gordon D, Garnaas K, Walton DS, Okjeldal OH, Guggenheim MA, Jackson LG, Elias ER, Moser HW (1995) Phenotype of patients with peroxisomal disorders subdivided into sixteen complementation groups. J Pediatr 127:13–22 [DOI] [PubMed] [Google Scholar]
- Moser HW (1999) Genotype-phenotype correlations in disorders of peroxisome biogenesis. Mol Genet Metab 68:316–327 [DOI] [PubMed] [Google Scholar]
- Portsteffen H, Beyer A, Becker E, Epplen C, Pawlak A, Kunau W-H, Dodt G (1997) Human PEX1 is mutated in complementation group 1 of the peroxisome biogenesis disorders. Nat Genet 17:449–452 [DOI] [PubMed] [Google Scholar]
- Reuber BE, Germain-Lee E, Collins CS, Morrell JC, Ameritunga R, Moser HW, Valle D, Gould SJ (1997) Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat Genet 17:445–448 [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Gould SJ (2000) The genetics of peroxisome biogenesis. Annu Rev Genet 34:623–652 [DOI] [PubMed] [Google Scholar]
- Singh I, Voigt RG, Sheikh FG, Kremser K, Brown FR III (1997) Biochemical features of a patient with Zellweger-like syndrome with normal PTS-1 and PTS-2 peroxisomal protein import systems: a new peroxisomal disease. Biochem Mol Med 61:198–207 [DOI] [PubMed] [Google Scholar]
- Slawecki ML, Dodt G, Steinberg S, Moser AB, Moser H, Gould SJ (1995) Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J Cell Sci 108:1817–1829 [DOI] [PubMed] [Google Scholar]
- Smeitink JA, Beemer FA, Espeel M, Donckerwolcke RA, Jakobs C, Wanders RJA, Schutgens RB, Roels F, Duran M, Dorland L, Berger R, Poll-The BT (1992) Bone dysplasia associated with phytanic acid accumulation and deficient plasmalogen synthesis: a peroxisomal entity amenable to plasmapheresis. J Inherit Metab Dis 15:377–380 [DOI] [PubMed] [Google Scholar]
- Tamura S, Okumoto K, Toyama R, Shimozawa N, Tsukamoto T, Suzuki Y, Osumi T, Kondo N, Fujiki Y (1998a) Human PEX1 cloned by functional complementation on a CHO cell mutant is responsible for peroxisome-deficient Zellweger syndrome of complementation group I. Proc Natl Acad Sci USA 95:4350–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Shimozawa N, Suzuki Y, Tsukamoto T, Osumi T, Fujiki Y (1998b) A cytoplasmic AAA family peroxin, Pex1p, interacts with Pex6p. Biochem Biophys Res Commun 245:883–886 [DOI] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RB, Wanders RJA, Tager JM (1992) Biochemistry of peroxisomes. Annu Rev Biochem 61:157–197 [DOI] [PubMed] [Google Scholar]
- Vreken P, van Lint AE, Bootsma AH, Overmars H, Wanders RJA, van Gennip AH (1998) Rapid stable isotope dilution analysis of very-long-chain fatty acids, pristanic acid and phytanic acid using gas chromatography-electron impact mass spectrometry. J Chromatogr B Biomed Sci Appl 713:281–287 [DOI] [PubMed] [Google Scholar]
- Wanders RJA (1999) Peroxisomal disorders: clinical, biochemical, and molecular aspects. Neurochem Res 24:565–580 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Mooijer PA, Dekker C, Suzuki Y, Shimozawa N (1999) Disorders of peroxisome biogenesis: complementation analysis shows genetic heterogeneity with strong overrepresentation of one group (PEX1 deficiency). J Inherit Metab Dis 22:314–318 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Ofman R, Romeijn GJ, Schutgens RB, Mooijer PA, Dekker C, van den Bosch H (1995) Measurement of dihydroxyacetone-phosphate acyltransferase (DHAPAT) in chorionic villous samples, blood cells and cultured cells. J Inherit Metab Dis 18:90–100 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Tager JM (1998) Lipid metabolism in peroxisomes in relation to human disease. Mol Aspects Med 19:69–154. [DOI] [PubMed] [Google Scholar]
- Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ (1998) Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet 63:347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PJ, Parniak MA, Akerman BR, Scriver CR (2000) Characterization of phenylketonuria missense substitutions, distant from the phenylalanine hydroxylase active site, illustrates a paradigm for mechanism and potential modulation of phenotype. Mol Genet Metab 69:101–110 [DOI] [PubMed] [Google Scholar]
- Waters PJ, Parniak MA, Hewson AS, Scriver CR (1998a) Alterations in protein aggregation and degradation due to mild and severe missense mutations (A104D, R157N) in the human phenylalanine hydroxylase gene (PAH). Hum Mutat 12:344–354 [DOI] [PubMed] [Google Scholar]
- Waters PJ, Parniak MA, Nowacki P, Scriver CR (1998b) In vitro expression analysis of mutations in phenylalanine hydroxylase: linking genotype to phenotype and structure to function. Hum Mutat 11:4–17 [DOI] [PubMed] [Google Scholar]
- Wei H, Kemp S, McGuinness MC, Moser AB, Smith KD (2000) Pharmacological induction of peroxisomes in peroxisome biogenesis disorders. Ann Neurol 47:286–296 [PubMed] [Google Scholar]
- Wiemer EA, Out M, Schelen A, Wanders RJ, Schutgens RB, Van den Bosch H, Tager JM (1991) Phenotypic heterogeneity in cultured skin fibroblasts from patients with disorders of peroxisome biogenesis belonging to the same complementation group. Biochim Biophys Acta 1097:232–237 [DOI] [PubMed] [Google Scholar]