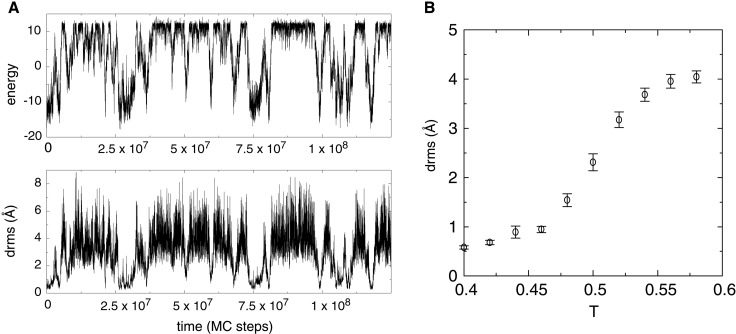
Figure 4.
Folding of the isolated helix 3 from the three-helix bundle protein. (A) A trajectory started from the native conformation of helix 3, run near the helix's transition temperature (T = 0.52). The same all-atom potential that was used to fold the entire protein is used here. Both energy and drms traces show that the helix repeatedly unfolds completely and refolds. (B) Average drms of helix 3 measured over long simulations at various temperatures. The average drms of fully unfolded conformations of the helix is ≈4 Å. Error bars indicate 1.5 SDs of the computed average.