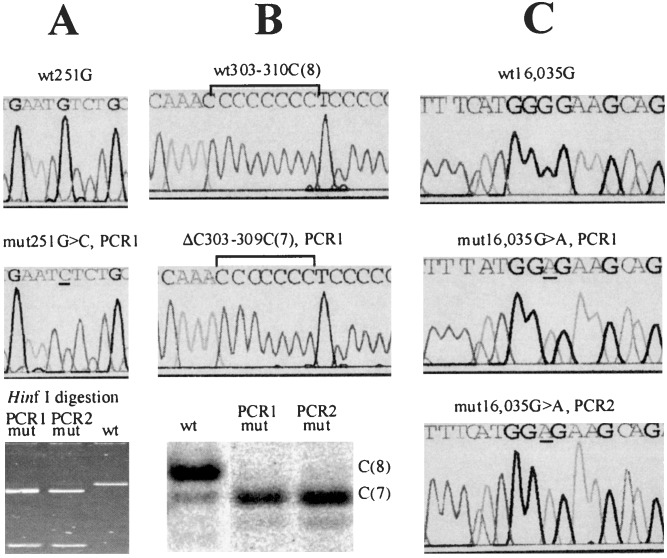
Figure 1.
Detection, quantification, and confirmation of point mutations in single cells. Somatic nucleotide changes are detected first by direct sequencing of PCR product from single cells. Mutations (Middle) are determined as changes in the sequence compared with the sequences from the majority of cells of the same individual (Top). After an initial identification of a mutation, a duplicate PCR was performed from the DNA of each presumably mutant cell (PCR 2). The mutations then were confirmed and quantified by one of the following methods, depending on the type of the mutation. Note that in each case, the two duplicate experiments yielded very similar results. mut, mutant; wt, wild type. (A) If the mutation resulted in a nucleotide change within a restriction site (in this example, a G → C change creates a HinfI restriction site, G′ANTC), direct restriction analysis was used to confirm and quantify the mutation by gel densitometry (Bottom). (B) A deletion/insertion within a C-tract was confirmed by a variant of the T-PCR approach (15). Briefly, a PCR fragment was restriction-digested to provide a labeled PCR/restriction fragment ≈50-nt long, which included the C-tract. The DNA was run on a sequencing gel. A deletion or insertion in the C-tract manifested itself as a band shift. The weaker subbands do not represent mutations but rather aberrant PCR products that ultimately limit the assay sensitivity. The image was quantified by using a PhosphorImager (Molecular Dynamics). (C) If a mutation did not alter a restriction site or the C-tract length, direct sequencing of the PCR fragments was used.