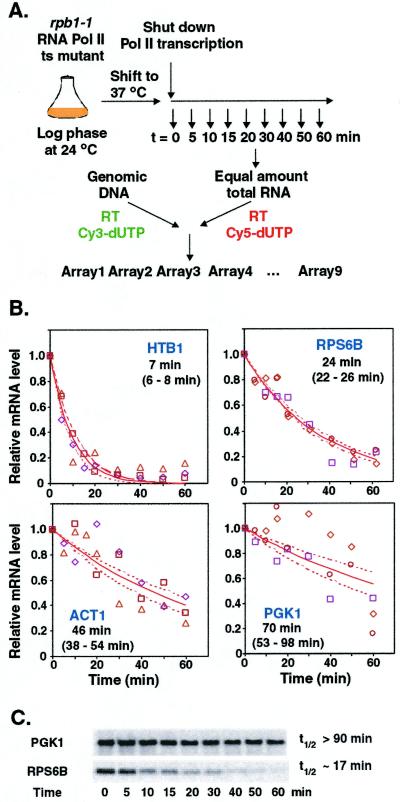
Figure 1.
Whole-genome determination of mRNA half-lives. (A) Schematic of the DNA microarray procedure for determining genomewide mRNA half-lives. Total RNA was isolated at specified intervals after inactivation of RNA polymerase II. Fluorescently labeled cDNA probes were prepared from each RNA sample by reverse transcription in the presence of Cy5-dUTP. Yeast genomic DNA was similarly labeled with Cy3-dUTP to provide an internal hybridization standard for every gene. For each time point, Cy5-labeled probe was mixed with the Cy3-labeled genomic DNA standard and the mixtures were hybridized to DNA microarrays. (B) Examples of mRNA decay profiles determined by quantitative microarray analysis. The different symbols represent data from three independent time courses; the solid lines represent the nonlinear least squares fit to an exponential decay model; the dashed lines represent the upper and lower limits of the 95% confidence interval for the decay curves, determined by the bootstrapping procedure. The calculated half-life and 95% confidence interval for the decay of each transcript are listed. (C) Northern analysis of the decay of mRNAs encoding PGK1 and RPS6B (quantified with a PhosphorImager).