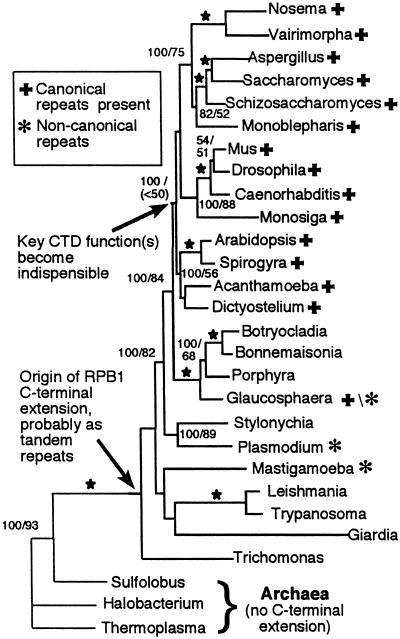
Figure 1.
Best tree and branch lengths recovered from maximum-likelihood analyses. Support values for each node from Bayesian inference and parsimony bootstrap analyses are shown either above or to the right of a given internode. In each case, Bayesian values are on the left of the slash with parsimony bootstrap values following. A star designates where both values are 100%. No values are given below 50% or for nodes that were not recovered in all three analyses. GenBank accessions used: Arabidopsis thaliana, P31635; Spirogyra sp., U90210; Bonnemaisonia hamifera, U90209; Botryocladia uvariodes, AF315819; Glaucosphaera vacuolata, AF315820; Porphyra yezoensis, U90208; Caenorhabditis elegans, P16356; Drosophila melanogaster, P04052; Monosiga brevicalis, AF315821; Mus musculus, NM_009089; Monoblepharis macandra, AF31582.2; Aspergillus oryzae, AB017184; Saccharomyces cerevisiae, P04050; Schizosaccharomyces pombe, P36594; Vairimorpha necatrix, AF060234; Nosema locustae, AF061288; Plasmodium falciparum, P14248; Stylonychia mytilus, AF315819; Acanthamoeba castellanii, U90211; Dictyostelium discoideum, AF58710/S52651; Mastigamoeba invertens, AF083338; Trichomonas vaginalis, U20501; Trypanosoma brucei, P17545; Leishmania donovani, AF126254; Halobacterium sp. NRC-1, AE005138; Sulfolobus acidocaldarius, X14818; Thermoplasma acidophilum, AL445064. The Giardia lamblia sequence was provided by H.-P. Klenk, Epidaurus, Bernried, Germany.