Abstract
Endothelial cells (EC) play a central role in inflammatory immune responses and efficiently induce effector functions in T cells, despite lacking the classical costimulatory ligands CD80 and CD86. By using the mAb HIL-131 we now demonstrate that human inducible costimulator-ligand (ICOS-L), a molecule related to CD80/CD86, is constitutively expressed on human EC in vivo. In vitro, ICOS-L expression was strongly enhanced on human umbilical vein EC and microvascular EC by the inflammatory cytokines tumor necrosis factor α and IL-1β, and to a lower extent by stimulation of EC by CD40 or lipopolysaccharide. Coculture of MHC class II+ EC with resting memory CD4+ T cells in the presence of superantigen led to a marked up-regulation of ICOS on T cells and to the production of Th1 (IFN-γ, IL-2) and Th2 cytokines (IL-4, IL-10, IL-13). When these cocultures were performed in the presence of the inhibitory mAb HIL-131, secretion of all cytokines was reduced by about 50–80%, indicating that ICOS-L is a major costimulator in EC-mediated T cell activation. Taken together, our data suggest an important physiological role of ICOS-L in the reactivation of effector/memory T cells on the endothelium controlling the entry of immune cells into inflamed tissue.
Inducible costimulator-ligand (ICOS-L) is a member of the B7 family of costimulatory ligands (1, 2) sharing 19–20% sequence identity with CD80 and CD86. Two splice variants of human ICOS-L have been described and designated hGL50 (3) and B7-H2/B7RP-1/hLICOS (4–6). Both molecules have an identical extracellular domain but differ at the carboxyl-terminal end of their cytoplasmic regions. In humans, cell surface expression of ICOS-L has been described on B cells, dendritic cells, monocytes/macrophages, and T cells (refs. 1 and 2; unpublished data). In addition, mRNA expression of ICOS-L has been detected in a variety of lymphoid and nonlymphoid organs, with hGL50 showing a more lymphoid-restricted expression pattern (spleen, lymph node), whereas B7-H2/B7RP-1/hLICOS mRNA was expressed in all organs examined (e.g., spleen, kidney, heart, and brain; ref. 7).
ICOS-L, unlike CD80 and CD86, does not interact with CD28 or CTLA-4 (CD152; refs. 8 and 9). Instead, ICOS-L binds to ICOS, a T cell-specific costimulatory molecule homologous to CD28 and CTLA-4 (8–10). The strong impact of ICOS/ICOS-L interaction on T cell-mediated immune responses in vivo became evident by the disruption of the ICOS gene in mice. ICOS-deficient mice are characterized by impaired germinal center formation, have a profound defect in isotype class switching in T cell-dependent B cell responses, and are defective in IL-4 and IL-13 production (11–13). In addition, blockade of ICOS/ICOS-L interaction in animal models of experimental allergic encephalomyelitis (14) and of cardiac allograft rejection (15) revealed a critical role of ICOS and its ligand in inflammatory immune reactions.
Endothelial cells (EC) play an important role in the recruitment of T cells and their activation at sites of inflammation (16–18). Human EC either constitutively express or up-regulate MHC class II molecules after exposure to IFN-γ (16–18). It has therefore been suggested that human EC might play a role in secondary immune responses by presenting antigen to circulating CD4+ memory T cells (18). Many in vitro studies have supported this notion by demonstrating that EC can stimulate CD4+ T cells in an antigen-specific manner (19, 20). However, in contrast to professional antigen-presenting cells like dendritic cells, human EC do not express the costimulatory ligands CD80 and CD86, which may explain the inability of EC to activate naive T cells (16, 18). Previous studies have demonstrated that activation of T cells by EC involves the interaction of LFA-3 (CD58) and OX40L (CD134L) with their corresponding receptors on T cells (21, 22). However, these molecules can account for only some of the accessory cell functions of EC, indicating the presence of an as-yet-unidentified costimulatory molecule.
In the present report, we demonstrate that human EC express ICOS-L, a member of the B-7 family of costimulatory ligands, in vitro and in vivo. Moreover, we describe its biochemical characteristics and its regulation by inflammatory stimuli, and we provide data demonstrating that ICOS-L plays an important functional role in the activation of T cells by EC.
Materials and Methods
Generation of L Cell Transfectants Expressing ICOS-L.
A human ICOS-L cDNA fragment was cloned into the expression vector BCMGSneo (23). The recombinant plasmid was transfected by electroporation into mouse fibroblastic Ltk− cells (L cells). Transfectants expressing ICOS-L were identified by flow cytometry by using a muICOS-Ig fusion protein (24).
Generation of ICOS-L-Specific mAbs.
mAbs directed against human ICOS-L were generated by immunizing BALB/c mice with a soluble human ICOS-L-rabbit-Ig fusion protein (9). Mouse spleen cells were fused with the myeloma P3X63Ag8.653 (American Type Culture Collection), and the resulting hybridomas were screened by flow cytometry with an ICOS-L-expressing L cell transfectant. After subcloning, 23 different mouse anti-ICOS-L mAbs were obtained and the antibody designated HIL-131 (IgG1) was used within this study.
Cell Isolation.
Human umbilical vein endothelial cells (HUVEC) were isolated from umbilical cords with 2.4 units/ml Dispase II (Roche Molecular Biochemicals) and cultured in endothelial growth medium with EC growth supplement (Promo Cell, Heidelberg) on gelatin-coated tissue culture vessels. Microvascular endothelial cells (MVEC) were obtained from explanted human hearts and cultured as described (25). Written consent was obtained from all donors; tissue sampling was approved by the local ethic committee. EC were used at passages 2–4 in all experiments. Cells were >97% CD144+, CD31+ and contained no contaminating CD14, CD16, CD19, CD3, and MHC class II expressing cells, as determined by flow cytometry. T cells were enriched from peripheral blood mononuclear cells by Ficoll–Hypaque gradient centrifugation and passage over a nylon-wool column. The enriched cells were then incubated with mAbs BU12 (anti-CD19; ref. 26), L243 (anti-MHC II), OKM1 (anti-CD11b), and OKT 8 (anti-CD8, all from American Type Culture Collection). CD4+CD45RA+ T cells were negatively selected by addition of mAb UCHL1 (anti-CD45RO; ref. 27) and CD4+CD45RO+ T cells were negatively selected by addition of mAb 4G11 (anti-CD45RA; ref. 28) by using goat anti-mouse IgG magnetobeads from Miltenyi Biotec, Bergisch-Gladbach, Germany. Purity was >95% as determined by flow cytometry with mAbs directed against CD14, CD16, CD3, CD4, CD40, CD45RO, and CD45RA.
Flow Cytometry.
Flow cytometry was performed on a FACSCalibur (BD Biosciences, Heidelberg) with the following mAbs either coupled to FITC, phycoerythrin (PE), or Cy5: B1.49.9 (anti-CD25, Immunotech, Marseille, France), F44 (anti-ICOS; ref. 10), HIL-131 (anti-ICOS-L), BK4 (anti-CD31, kindly provided by B. Pötsch, Bad Nauheim, Germany), BU12 (anti-CD19), 91d6 (anti-CD4; ref. 29), UCHL1 (anti-CD45RO), B73.1 (anti-CD16; ref. 30), 63D3 (anti-CD14), OKT 3 (anti-CD3), L243 (anti-MHC II), G28.5 (anti-CD40), 1H3 (anti-CD62L; all from American Type Culture Collection), G43–25B (anti-CD11a), 1G10 (anti-CD43), G44–26 (anti-CD44), KPL-1 (anti-CD162), 1C6/CXCR3 (anti-CXCR3), 12G5 (anti-CXCR4), 2H4 (anti-CCR7), HECA-452 (anti-CLA; all from BD PharMingen), BU49 (anti-CD49d), BRIC-5 (anti-CD58), BV6 (anti-CD144; all from Chemicon). In addition to HIL-131, a soluble human ICOS-rabbit-Ig fusion protein coupled to PE was used for ICOS-L staining. mAb 2A11 (IgG1, own hybridoma) and CD3δ-rabbit-Ig coupled to the appropriate chromophore, were used as isotype controls. The specificity of binding of HIL-131 and ICOS-Ig was further controlled by blocking experiments with a 50- to 100-fold excess of an unconjugated reagent (cold blocking control).
Reverse Transcription–PCR of ICOS-L Splice Variants.
RNA obtained from untreated or tumor necrosis factor α (TNF-α)-stimulated (48 h) HUVEC was reverse-transcribed by using the Superscript II enzyme from Life Technologies, Rockville, MD. The single-stranded cDNA was used to amplify the ICOS-L splice variants hGL50 and B7-H2/B7RP-1/hLICOS by PCR with Pfu DNA polymerase from Stratagene. The forward primer VL141 and the downstream primers VL274 (specific for B7-H2/B7RP-1/hLICOS) and VL162B (specific for hGL50) used for amplification have been described (7). Primers VL274 and VL162B were slightly modified by the addition of a NotI restriction site to their 3′ end. Cycle conditions were 94°C for 40 sec, 62°C for 1 min, and 75°C for 2 min for 38 cycles. The PCR products were separated on 2% agarose gels.
Immunoprecipitation.
HUVEC (15 × 106 cells) were stimulated for 24 h with 200 units of TNF-α and cell surface iodinated with 0.75 mCi 125I by using IODO-Beads (Pierce). HUVEC were lysed in a Nonidet P-40 buffer and ICOS-L was immunoprecipitated by using mAb HIL-131 covalently coupled to protein G-Sepharose (Amersham Pharmacia; ref. 10). The immunoprecipitates were separated by reducing or nonreducing SDS/PAGE (10% gel). To determine the molecular mass (Mr) of the polypeptide backbone of ICOS-L, immunoprecipitates were treated with 8 units of N-glycosidase F (Roche Molecular Biochemicals).
Immunohistology.
Cryostat sections (8 μm) of umbilical cords or human tonsils were prepared, fixed in acetone, and immunostained with the mAb HIL-131 or BV6 (anti-CD144), followed by biotinylated goat anti-mouse antibody (Dianova, Hamburg, Germany) and streptavidin-horseradish peroxidase (Dianova); 3-amino-9-ethylcarbazole was used as substrate (Sigma). Nuclei were counterstained with hematoxylin.
Stimulation of HUVEC and MVEC.
Cells (7 × 105) were seeded into 6-cm Petri dishes and treated with either 1,000 units/ml IFN-γ (BioSource International, Camarillo, CA), 200 units/ml TNF-α (R&D Systems), 10 ng/ml IL-1β (Strathmann Biotech, Hannover, Germany), 1 μg/ml lipopolysaccharide (LPS) (Sigma), or 400 units/ml IL-4 (Novartis, Vienna) for 24 h. EC were also stimulated by CD40 for 24 h with CD40L-expressing, paraformaldehyde-fixed P3xTB.A7-transfectants (31). P3X63Ag8.653 wild-type cells were used as a control. Ratio of EC to P3X63Ag8.653 was 1:5. After stimulation, cells were analyzed for ICOS-L expression by flow cytometry.
Coculture of HUVEC with T Cells.
HUVEC (7 × 104) were seeded into 24-well plates and stimulated with 1,000 units/ml IFN-γ for 24–48 h to induce MHC class II expression. The cells were washed, and 7 × 105 CD4+CD45RA+ or CD4+CD45RO+ T cells were added in the presence of a mixture of staphylococcal enterotoxin B, staphylococcal enterotoxin A, and toxic shock syndrome toxin-1 (10 pg/ml each; all from Toxin Technology, Sarasota, FL). After indicated times, the T cells were removed and analyzed for the expression of ICOS and CD25 by flow cytometry. In separate experiments we have also analyzed the expression of T cell adhesion molecules in the absence or presence of the blocking mAb HIL-131 (15 μg/ml). For measurement of cytokine production, CD4+CD45RA+ or CD4+CD45RO+ T cells were cocultured with HUVEC (pretreated either with 1,000 units/ml IFN-γ alone or in some experiments with 1,000 units/ml IFN-γ and 200 units/ml TNF-α) as described above in the presence of superantigen (SAg) (1.5 pg/ml staphylococcal enterotoxin B, staphylococcal enterotoxin A, toxic shock syndrome toxin-1 each). Where indicated, TS2/9 (5 μg/ml, anti-LFA-3, Endogen, Cambridge, MA), HIL-131 (anti-ICOS-L), L243 (anti-MHC II), or 2A11 (isotype control; 15 μg/ml each) were added. The supernatants were collected after 36 h and assayed for cytokine secretion by ELISA.
Cytokine Measurements.
IFN-γ was determined by ELISA by using mAbs 4SB3 and 7R2A4 (European Collection of Animal Cell Culture, Salisbury, U.K.; refs. 32 and 33). Commercial ELISA kits were used to measure IL-2, IL-4, and IL-10 (BioSource International), and IL-13 (BD PharMingen). The amount of cytokines produced varied between individual experiments (IL-4, 0–256 pg/ml; IL-10, 0–2,614 pg/ml; IL-13, 1,200–2,797 pg/ml; IFN-γ, 570–7,525 pg/ml, IL-2, 442–10,850 pg/ml).
Carboxyfluorescein Diacetate Succinimidyl Ester (CSFE) Labeling and Cell Division Analysis.
CD4+CD45RA+ or CD4+CD45RO+ T cells were labeled with 0.5 μM CSFE (Molecular Probes) for 10 min at room temperature and cocultured with IFN-γ-pretreated HUVEC as described above in the presence of SAg (10 pg/ml staphylococcal enterotoxin B, staphylococcal enterotoxin A, toxic shock syndrome toxin-1 each). Where indicated, 15 μg/ml of the following reagents were used: HIL-131 (anti-ICOS-L), L243 (anti-MHC II), or 2A11 (isotype control). After 5 days, T cells were analyzed for cell division by flow cytometry.
Results
mAb HIL-131 Binds to ICOS-L and Inhibits the Interaction with ICOS.
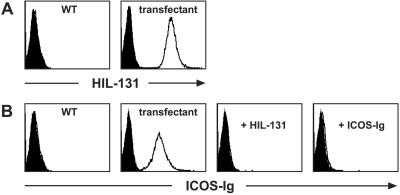
To analyze ICOS-L expression and function we generated a panel of mAbs directed to human ICOS-L. The specificity of these mAbs was confirmed by staining of ICOS-L transfectants. The reactivity pattern of one of the mAbs obtained (clone HIL-131) is shown in Fig. 1A. We further investigated the capacity of our mAbs to prevent binding of ICOS to its ligand. We found that mAb HIL-131, at low concentrations, fully blocked the binding of a human ICOS-Ig fusion protein to ICOS-L-transfected L cells (Fig. 1B). To assess the blocking efficiency of HIL-131, the blockade obtained with an excess of unconjugated ICOS-Ig fusion protein is shown for comparison (Fig. 1B).
Figure 1.
Characterization of the ICOS-L mAb HIL-131. (A) Binding specificity of HIL-131. Wild-type and ICOS-L-transfected L cells were stained with PE-labeled mAb HIL-131 (open profiles) or an isotype control (filled profiles). (B) Blocking capacity of HIL-131. Wild-type and ICOS-L-transfected L cells were stained with a PE-coupled human ICOS-rabbit-Ig fusion protein (2 μg/ml, open profiles) or a control fusion protein (CD3δ-rabbit-Ig-PE, filled profiles). Staining was blocked by preincubation of the cells with either mAb HIL-131 (10 μg/ml) or an excess of unlabeled human ICOS-rabbit-Ig (100 μg/ml). WT, wild type.
ICOS-L Is Constitutively Expressed by EC and Strongly Up-Regulated by Inflammatory Stimuli.
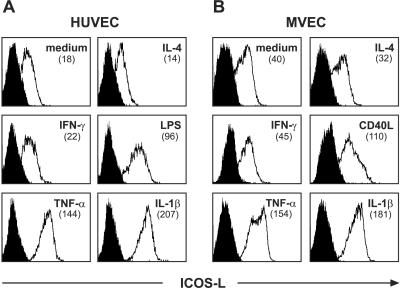
To explore the expression of ICOS-L on EC, we stained isolated HUVEC and MVEC with the ICOS-L-specific mAb HIL-131. As shown in Fig. 2 A and B, both cell types constitutively express ICOS-L on their cell surface. A similar staining was observed with a different anti-ICOS-L mAb and a soluble ICOS-Ig fusion protein (data not shown). Recently, it has been demonstrated that ICOS-L expression could be induced in murine fibroblasts (34) and in CD34+ hematopoietic progenitor cells (35) by TNF-α. We therefore investigated the influence of TNF-α and other inflammatory stimuli (IL-1β, IFN-γ, IL-4, LPS, and CD40-triggering) on ICOS-L expression in MVEC and/or HUVEC by flow cytometry. Whereas IFN-γ and IL-4 were without effect (Fig. 2 A and B), LPS (Fig. 2A) or stimulation of EC by CD40 (Fig. 2B) led to an increase in ICOS-L expression. The most striking effect was observed with the inflammatory cytokines IL-1β and TNF-α, which both very effectively enhanced ICOS-L expression on HUVEC and MVEC (Fig. 2 A and B). In clear contrast to the expression of ICOS-L by EC, expression of CD80 or CD86 could not be detected on HUVEC or MVEC (data not shown; refs. 16 and 18).
Figure 2.
Expression of ICOS-L by EC. HUVEC (A) or MVEC (B) were left untreated or were treated with the indicated reagents or with the CD40L transfectant P3xTB.A7 for 24 h, and ICOS-L expression was measured by flow cytometry by using PE-conjugated mAb HIL-131. Filled profiles represent cold blocking controls. In contrast to the CD40L transfectant, wild-type cells did not influence ICOS-L expression (data not shown). The numbers in brackets indicate the mean fluorescence intensity. One representative experiment of three is shown.
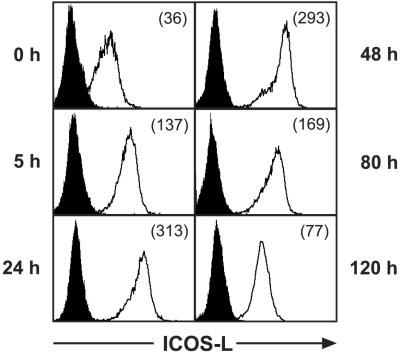
Time Course of ICOS-L Expression by TNF-α-Treated HUVEC.
HUVEC were stimulated with TNF-α, and ICOS-L expression was investigated after various times by flow cytometry. ICOS-L up-regulation was already detectable 5 h after stimulation, reached a maximum at 24–48 h, and gradually declined thereafter (Fig. 3). The continuos presence of TNF-α seemed to be necessary for high-level ICOS-L expression, because removal of TNF-α after 24 h by washing the cells and culturing them in fresh medium led to a rapid decrease of ICOS-L expression (data not shown).
Figure 3.
Kinetics of cell surface ICOS-L expression. HUVEC were stimulated with 200 units/ml TNF-α for the times indicated and analyzed for ICOS-L expression by flow cytometry with PE-conjugated mAb HIL-131. Filled profiles represent blocking controls. The numbers in brackets indicate the mean fluorescence intensity. One representative experiment of three is shown.
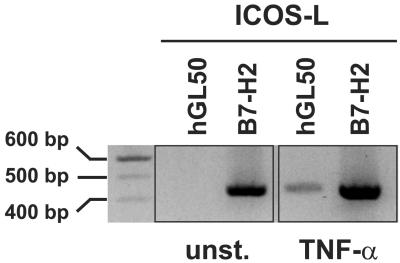
Both ICOS-L Splice Variants Are Expressed by EC.
To analyze the expression of ICOS-L splice variants in EC, we performed reverse transcription–PCR with primer sets specific for hGL50 and B7-H2/B7RP-1/hLICOS by using RNA isolated from unstimulated and TNF-α-stimulated HUVEC. As shown in Fig. 4, transcripts for B7-H2/B7RP-1/hLICOS could be detected in unstimulated and stimulated HUVEC, whereas a signal for hGL50 was observed only with RNA obtained from TNF-α-treated HUVEC. These results indicate that both ICOS-L splice variants are made by EC.
Figure 4.
Reverse transcription–PCR analysis of ICOS-L splice variants. cDNA generated from unstimulated or TNF-α-stimulated HUVEC was amplified by using primer pairs specific for hGL50 or B7-H2/B7RP-1/hLICOS. The resulting PCR fragments were separated by agarose gel electrophoresis. Twenty microliters of the hGL50 and 10 μl of the B7-H2/B7RP-1/hLICOS PCR products were loaded. Unst., unstimulated.
Biochemical Characterization of ICOS-L Expressed by EC.
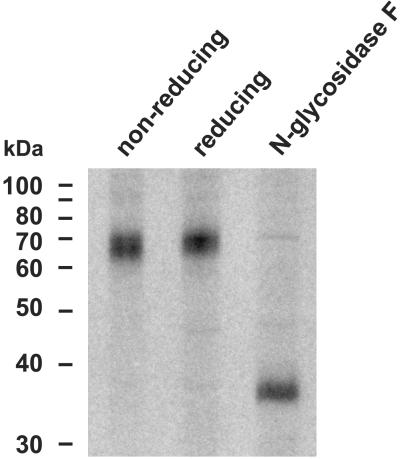
To determine the biochemical features of ICOS-L, we performed immunoprecipitation studies on TNF-α-stimulated HUVEC by using mAb HIL-131. We detected one single band at 63–72 kDa under nonreducing and reducing conditions (Fig. 5), indicating that ICOS-L is expressed as a monomer on the surface of EC. Treatment of the immunoprecipitate with N-glycosidase F reduced the apparent Mr of ICOS-L to approximately 35–38 kDa (Fig. 5), indicating that ICOS-L is highly glycosylated. The Mr of the deglycosylated protein slightly differs from the predicted Mr of the mature protein backbone of ICOS-L (31.4 kDa for B7-H2/B7RP-1/hLICOS or 32.3 kDa for hGL50 without the putative signal peptide), suggesting other as-yet-undefined posttranslational modifications.
Figure 5.
Immunoprecipitation of ICOS-L. TNF-α-stimulated HUVEC were surface-iodinated and ICOS-L protein was immunoprecipitated with anti-ICOS-L mAb HIL-131. Immunoprecipitates were separated by SDS/PAGE under nonreducing or reducing conditions. In some experiments, the immunoprecipitate was treated by N-glycosidase F before analysis under reducing conditions.
ICOS-L Expression by EC in Vivo.
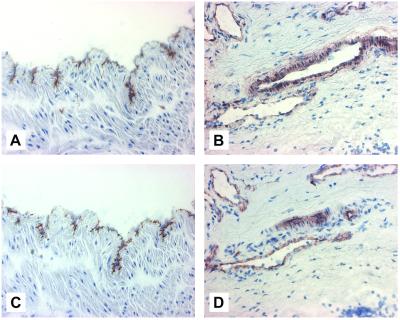
To support our in vitro findings, we examined the expression of ICOS-L in vivo by immunostaining frozen tissue sections from umbilical cords and human tonsils with mAb HIL-131. The venular (Fig. 6A) and the arteriolar (data not shown) EC layers of the umbilical cord stained strongly for ICOS-L. ICOS-L expression on EC was also observed in other tissues, as exemplified by the staining of small blood vessel EC in human tonsils (Fig. 6B). These data demonstrate that ICOS-L is expressed on EC in vivo. For comparison, also shown is the staining pattern of VE-Cadherin (CD144), an EC-specific marker (ref. 36; Fig. 6 C and D).
Figure 6.
In vivo analysis of ICOS-L expression by immunohistochemistry. Frozen tissue sections of umbilical cords (A and C) and human tonsils (B and D) were immunostained with the ICOS-L-specific mAb HIL-131 (A and B) or with the EC-specific mAb BV6 (anti-CD144) for comparison (C and D).
EC-Induced Up-Regulation of ICOS on T Cells.
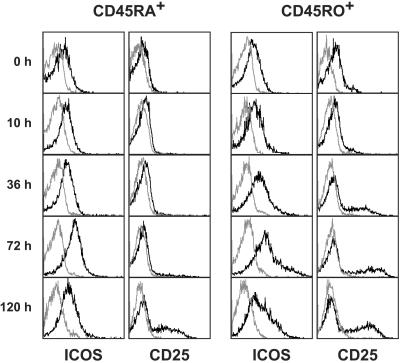
To investigate whether EC have the capacity to induce ICOS expression on T cells, we cocultured CD4+CD45RA+ or CD4+CD45RO+ T cells with IFN-γ-stimulated HUVEC in the presence of SAg and examined the expression of ICOS at various time points by flow cytometry. Purified CD4+CD45RA+ T cells alone exhibited a low basal expression of ICOS (Fig. 7). On activation of these cells with HUVEC, a slight increase in ICOS expression was observed with a maximum at 72 h. When HUVEC were cocultured with CD4+CD45RO+ T cells, ICOS was more rapidly and more strongly induced (mean fluorescence intensity, 44 vs. 190 at 72 h; Fig. 7). Up-regulation of ICOS was not observed on T cells stimulated with SAg alone at any time point (data not shown). For comparison also shown is the expression of CD25 (Fig. 7).
Figure 7.
Cell surface staining of EC-activated T cells. INFγ-activated HUVEC were cocultured with CD4+CD45RA+ or CD4+CD45RO+ T cells in the presence of SAg. After the indicated times, T cells were analyzed for cell surface expression of ICOS (F44-PE) and CD25 (B1.49.9-FITC) by flow cytometry. Gray profiles represent isotype controls. Expression analyses with naive T cells were performed twice and with memory T cells five times.
ICOS-L mAb HIL-131 Inhibits EC-Mediated Cytokine Production But Not the Expression of T Cell Adhesion Molecules.
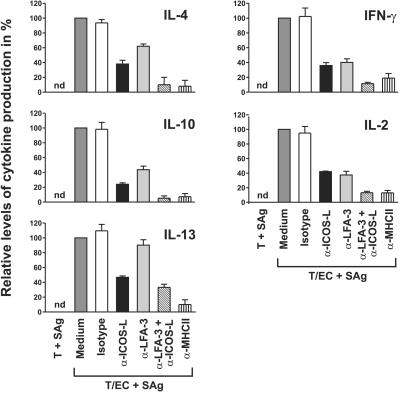
We next investigated the functional consequences of the interaction of ICOS-L on EC with ICOS on T cells. To this end, MHC class II+ HUVEC were cocultured with CD4+CD45RA+ or CD4+CD45RO+ T cells and SAg in the absence or presence of the inhibitory mAb HIL-131, and production of Th1 (IFN-γ, IL-2) and Th2 cytokines (IL-4, IL-10, IL-13) was measured by ELISA. In agreement with previously published results, cytokine production could be observed only when HUVEC were cocultured with memory, but not with naive T cells (irrespective of whether the EC have been pretreated with IFN-γ alone or with INF-γ and TNF-α; data not shown and ref. 37). In cocultures with CD4+CD45RO+ T cells, HIL-131 inhibited the production of all cytokines by about 50–80% (Fig. 8). A similar result was obtained with a different mAb directed against ICOS-L (data not shown). For comparison, cocultures were also performed in the presence of TS2/9 (anti-LFA-3), a mAb known to inhibit T cell activation by EC (21). As a result, TS2/9 blocked the secretion of IL-2 and IFN-γ to a similar extent as HIL-131, and also inhibited the secretion of IL-4 and IL-10, albeit with lower efficiency. In contrast to mAb HIL-131, mAb TS2/9 did not inhibit IL-13 production (Fig. 8). By combining the blocking effects achieved with HIL-131 and TS2/9, the inhibition reached levels obtained with the anti-MHC class II mAb L243 (Fig. 8). No cytokines could be detected, when T cells were cultured with SAg in the absence of EC.
Figure 8.
Inhibition of cytokine production in EC/T cell cocultures by blocking ICOS/ICOS-L interaction. INFγ-activated HUVEC and CD4+CD45RO+ T cells were cocultured with SAg in the absence or presence of an isotype control mAb (2A11), anti-ICOS-L mAb (HIL-131), anti-LFA-3 mAb (TS2/9), anti-ICOS-L mAb + anti-LFA-3 mAb, or anti-MHC class II mAb (L243). Cell culture supernatants were assayed for cytokine secretion after 36 h by ELISA. Secretion of IL-4 and IL-10 was donor-dependent and could not be measured in all of the experiments performed. Cytokine production in the medium control was set to 100%. Error bars represent the means ± SE of 4–6 experiments. T, CD4+CD45RO+ T cells; nd, not detectable.
In separate EC/T cell coculture experiments we also analyzed the influence of ICOS/ICOS-L interaction on the expression of various T cell adhesion molecules (CD62L, CD162, CLA, CD49d, CD11a, LFA-3, CXCR3, CXCR4, CCR7, CD43, and CD44) by CD45RO+ T cells. As a result, we found that the expression levels of none of these markers could be influenced by the blocking mAb HIL-131 (data not shown).
Influence of ICOS/ICOS-L Interaction on EC-Induced T Cell Proliferation.
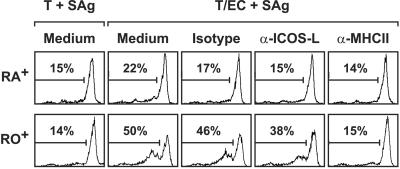
Carboxyfluorescein diacetate succinimidyl ester-labeled CD4+CD45RA+ or CD4+CD45RO+ T cells were cocultured with MHC class II+ HUVEC and SAg in the absence or presence of mAb directed to ICOS-L, or MHC class II, or an isotype control mAb and analyzed for cell division by flow cytometry. As expected, an EC-driven proliferation of CD4+CD45RA+ T cells could hardly be detected (Fig. 9). In contrast, when cocultures were performed with CD4+CD45RO+ T cells, up to 50% of the T cells divided. Whereas treatment of the cocultures with the anti-MHC class II mAb L243 reduced T cell proliferation to background levels, only a slight inhibition was observed by treatment of the cocultures with HIL-131, indicating that ICOS/ICOS-L-interaction has only a minor influence on EC-induced T cell proliferation (Fig. 9).
Figure 9.
Influence of EC-expressed ICOS-L on T cell proliferation. CD4+CD45RA+ or CD4+CD45RO+ T cells were labeled with carboxyfluorescein diacetate succinimidyl ester and cocultured with IFN-γ-stimulated HUVEC and SAg for 5 days in the absence or presence of an isotype control mAb (2A11), anti-ICOS-L mAb (HIL-131), or anti-MHC class II mAb (L243). T cells were analyzed for cell division by flow cytometry. Data are representative of two experiments. T, T cells.
Discussion
The expression of costimulatory ligands of the B7 family enables antigen-presenting cells to initiate and maintain effective T cell responses (1, 2). Although human EC do not express CD80 or CD86, they nevertheless have the capacity to potently activate T cell effector functions in vitro.
We now demonstrate that human EC are endowed with a “professional” costimulatory ligand of the B7 family, ICOS-L. We further show that ICOS-L, a glycosylated monomer of 63–72 kDa, is expressed on the cell surface of EC in vitro and in vivo, and is up-regulated by various inflammatory stimuli including TNF-α, IL-1β, and LPS, or by triggering by CD40. Most importantly, ICOS-L on EC is functional, as demonstrated by its capacity to costimulate cytokine secretion in cocultures of T cells with EC. Together with the observation that EC express MHC class II molecules, the finding of ICOS-L expression on EC in vivo lends strong support to the notion that EC function as accessory cells for CD4+ memory T cells in vivo.
The strong impact of ICOS/ICOS-L interaction in EC-mediated T-cell activation became evident by direct comparison with the CD2/LFA-3 system. Blocking the interaction of ICOS with its ligand during EC/T cell cocultures inhibited cytokine secretion to the same extent (IL-2, IFN-γ) or even more efficiently (IL-4, IL-10, IL-13) when compared with the blockade of the CD2/LFA-3 receptor ligand pair. Triggering of ICOS by ICOS-L expressed by EC had a profound effect on the secretion of Th1 as well as Th2 cytokines, a finding that corroborates the hypothesis that ICOS/ICOS-L does not only have a role in the control of Th2, but might also have a critical function in the control of Th1 immune responses (38). On the basis of previous data, we and others suggested a model in which CD28 costimulates primary T cell functions, whereas ICOS plays a more prominent role in the costimulation of effector or memory T cell responses (10, 39). Our functional data presented here are in agreement with such a concept. In contrast to SAg-driven cocultures performed with EC and memory T cells, cocultures with naive CD4+CD45RA+ T cells did not result in any measurable cytokine secretion (ref. 37 and this report), a finding that may be attributed to the lack of CD80/CD86 expression on EC. Thus, our data clearly demonstrate that ICOS-L expressed on EC delivers costimulatory signals to memory cells, but cannot replace CD80/CD86 in priming naive T cells.
As revealed by mRNA expression studies, ICOS-L is expressed in lymphoid and nonlymphoid organs (3, 7). B cells, T cells, and dendritic cells, which all have been shown to express ICOS-L on their cell surface, may account for ICOS-L expression in lymphoid organs; the cell type responsible for high-level expression of ICOS-L in nonlymphoid tissues, e.g., kidney, heart, or brain, has not been identified to date. Our data indicate that EC represent a major source of ICOS-L in these organs. Expression of ICOS-L on EC may also explain findings from a previous study in mice, in which a rapid up-regulation of ICOS-L was observed in various nonlymphoid tissues after in vivo injection of LPS (34). Moreover, both ICOS-L splice variants are expressed by EC stimulated with TNF-α, indicating that under certain inflammatory conditions ICOS-L splice variant hGL50 might also be expressed on nonlymphoid tissues, in addition to B7-H2/B7RP-1/hLICOS. With regard to tissue distribution, ICOS-L differs strikingly from CD80 and CD86, whose expression is largely confined to professional antigen-presenting cells within lymphoid tissue (2, 34, 40), suggesting a unique function for ICOS-L during immune reactions in nonlymphoid organs.
EC and CD4+ T cells interact and are activated during immune reactions and, if not properly controlled, are thought to contribute to the immunopathogenesis of inflammatory diseases. In this context, our finding that the basal expression of ICOS-L on EC could be strongly enhanced by inflammatory mediators like TNF-α, IL-1β, or LPS, is of particular interest. On the basis of these findings, it can be assumed that in the course of an infection, cells of the innate immune system will locally produce the inflammatory cytokines IFN-γ, TNF-α, and IL-1. Stimulation by IFN-γ will enable EC to present antigen by up-regulating MHC-class II molecules, whereas stimulation by TNF-α/IL-1 will endow EC with the ICOS-L. ICOS-expressing antigen-specific T cells, generated in secondary lymphoid organs, once attracted to the site of inflammation, will reencounter their cognate antigen presented by EC. Costimulation of these effector T cells by ICOS is likely to significantly contribute to their reactivation and subsequent extravasation to the underlying tissue. A similar mechanism may be operative in noninfectious inflammatory conditions. For example, ICOS/ICOS-L-interaction was critical in experimental allergic encephalomyelitis and allograft rejection (14, 15), murine disease models in which CD4+ T cells and EC are known to participate in the disease process.
Collectively, our data suggest that ICOS-L expressed on EC, in the absence of CD80 and CD86, plays a major role for the reactivation of effector/memory T cells on the endothelium controlling the entry of immune cells into inflamed tissue.
Acknowledgments
The expert technical assistance of Jenny Ehlert is gratefully acknowledged. The study was supported by Grants Ma 1912/2–1 (to H.W.M.) and SFB 506 (to R.A.K.) from the Deutsche Forschungsgemeinschaft.
Abbreviations
- EC
endothelial cells
- ICOS-L
inducible costimulator-ligand
- HUVEC
human umbilical vein endothelial cells
- MVEC
microvascular endothelial cells
- LPS
lipopolysaccharide
- SAg
superantigen
- TNF-α
tumor necrosis factor α
- PE
phycoerythrin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Coyle A J, Gutierrez-Ramos J C. Nat Immunol. 2001;2:203–209. doi: 10.1038/85251. [DOI] [PubMed] [Google Scholar]
- 2.Chambers CA. Trends Immunol. 2001;22:217–223. doi: 10.1016/s1471-4906(01)01868-3. [DOI] [PubMed] [Google Scholar]
- 3.Ling V, Wu P W, Finnerty H F, Bean K M, Spaulding V, Fouser L A, Leonard J P, Hunter S E, Zollner R, Thomas J L, et al. J Immunol. 2000;164:1653–1657. doi: 10.4049/jimmunol.164.4.1653. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Zhu G, Chapoval A I, Dong H, Tamada K, Ni J, Chen L. Blood. 2000;96:2808–2813. [PubMed] [Google Scholar]
- 5.Yoshinaga S K, Zhang M, Pistillo J, Horan T, Khare S D, Miner K, Sonnenberg M, Boone T, Brankow D, Dai T, et al. Int Immunol. 2000;12:1439–1447. doi: 10.1093/intimm/12.10.1439. [DOI] [PubMed] [Google Scholar]
- 6.Brodie D, Collins A V, Iaboni A, Fennelly J A, Sparks L M, Xu X N, van der Merwe P A, Davis S J. Curr Biol. 2000;10:333–336. doi: 10.1016/s0960-9822(00)00383-3. [DOI] [PubMed] [Google Scholar]
- 7.Ling V, Wu P W, Miyashiro J S, Marusic S, Finnerty H F, Collins M. J Immunol. 2001;166:7300–7308. doi: 10.4049/jimmunol.166.12.7300. [DOI] [PubMed] [Google Scholar]
- 8.Yoshinaga S K, Whoriskey J S, Khare S D, Sarmiento U, Guo J, Horan T, Shih G, Zhang M, Coccia M A, Kohno T, et al. Nature (London) 1999;402:827–832. doi: 10.1038/45582. [DOI] [PubMed] [Google Scholar]
- 9.Beier K C, Hutloff A, Dittrich A M, Heuck C, Rauch A, Büchner K, Ludewig B, Ochs H D, Mages H W, Kroczek R A. Eur J Immunol. 2000;30:3707–3717. doi: 10.1002/1521-4141(200012)30:12<3707::AID-IMMU3707>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Hutloff A, Dittrich A M, Beier K C, Eljaschewitsch B, Kraft R, Anagnostopoulos I, Kroczek R A. Nature (London) 1999;397:263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 11.Dong C, Juedes A E, Temann U A, Shresta S, Allison J P, Ruddle N H, Flavell R A. Nature (London) 2001;409:97–101. doi: 10.1038/35051100. [DOI] [PubMed] [Google Scholar]
- 12.McAdam A J, Greenwald R J, Levin M A, Chernova T, Malenkovich N, Ling V, Freeman G J, Sharpe A H. Nature (London) 2001;409:102–105. doi: 10.1038/35051107. [DOI] [PubMed] [Google Scholar]
- 13.Tafuri A, Shahinian A, Bladt F, Yoshinaga S K, Jordana M, Wakeham A, Boucher L M, Bouchard D, Chan V S, Duncan G, et al. Nature (London) 2001;409:105–109. doi: 10.1038/35051113. [DOI] [PubMed] [Google Scholar]
- 14.Rottman J B, Smith T, Tonra J R, Ganley K, Bloom T, Silva R, Pierce B, Gutierrez-Ramos J C, Özkaynak E, Coyle A J. Nat Immunol. 2001;2:605–611. doi: 10.1038/89750. [DOI] [PubMed] [Google Scholar]
- 15.Özkaynak E, Gao W, Shemmeri N, Wang C, Gutierrez-Ramos J C, Amaral J, Qin S, Rottman J B, Coyle A J, Hancock W W. Nat Immunol. 2001;2:591–596. doi: 10.1038/89731. [DOI] [PubMed] [Google Scholar]
- 16.Rose M L. Cell Mol Life Sci. 1998;54:965–978. doi: 10.1007/s000180050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cines D B, Pollak E S, Buck C A, Loscalzo J, Zimmerman G A, McEver R P, Pober J S, Wick T M, Konkle B A, Schwartz B S, et al. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- 18.Pober JS. Immunol Res. 1999;19:225–232. doi: 10.1007/BF02786490. [DOI] [PubMed] [Google Scholar]
- 19.Vora M, Yssel H, de Vries J E, Karasek M A. J Immunol. 1994;152:5734–5741. [PubMed] [Google Scholar]
- 20.Savage C O, Brooks C J, Harcourt G C, Picard J K, King W, Sansom D M, Willcox N. Int Immunol. 1995;7:471–479. doi: 10.1093/intimm/7.3.471. [DOI] [PubMed] [Google Scholar]
- 21.Hughes C C, Savage C O, Pober J S. J Exp Med. 1990;171:1453–1467. doi: 10.1084/jem.171.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunitomi A, Hori T, Imura A, Uchiyama T. J Leukocyte Biol. 2000;68:111–118. [PubMed] [Google Scholar]
- 23.Karasuyama H, Tohyama N, Tada T. J Exp Med. 1989;169:13–25. doi: 10.1084/jem.169.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mages H W, Hutloff A, Heuck C, Büchner K, Himmelbauer H, Oliveri F, Kroczek R A. Eur J Immunol. 2000;30:1040–1047. doi: 10.1002/(SICI)1521-4141(200004)30:4<1040::AID-IMMU1040>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Gräfe M, Auch-Schwelk W, Graf K, Terbeek D, Hertel H, Unkelbach M, Hildebrandt A, Fleck E. Am J Physiol. 1994;267:H2138–H2148. doi: 10.1152/ajpheart.1994.267.6.H2138. [DOI] [PubMed] [Google Scholar]
- 26.Flavell D J, Flavell S U, Boehm D A, Emery L, Noss A, Ling N R, Richardson P R, Hardie D, Wright D H. Br J Cancer. 1995;72:1373–1379. doi: 10.1038/bjc.1995.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith S H, Brown M H, Rowe D, Callard R E, Beverley P C. Immunology. 1986;58:63–70. [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto C. In: Leucocyte Typing V. White Cell Differentiation Antigens. Schlossman S F, Boumsell L, Gilks W, Harlan J M, Kishimoto T, Morimoto C, Ritz J, Shaw S, Silverstein R, Springer T, et al., editors. Oxford: Oxford Univ. Press; 1995. pp. 386–389. [Google Scholar]
- 29.Gramatzki M, Baum W, Burmester G R, Kalden J R. Eur J Immunol. 1984;14:762–765. doi: 10.1002/eji.1830140819. [DOI] [PubMed] [Google Scholar]
- 30.Perussia B, Starr S, Abraham S, Fanning V, Trinchieri G. J Immunol. 1983;130:2133–2141. [PubMed] [Google Scholar]
- 31.Graf D, Müller S, Korthäuer U, van Kooten C, Weise C, Kroczek R A. Eur J Immunol. 1995;25:1749–1754. doi: 10.1002/eji.1830250639. [DOI] [PubMed] [Google Scholar]
- 32.Meager A, Parti S, Barwick S, Spragg J, O'Hagan K. J Interferon Res. 1984;4:619–625. doi: 10.1089/jir.1984.4.619. [DOI] [PubMed] [Google Scholar]
- 33.Meager A. In: Lymphokines and Interferon: A Practical Approach. Clemens M J, Morris A G, Gearing A J H, editors. Oxford: IRL; 1987. pp. 105–127. [Google Scholar]
- 34.Swallow M M, Wallin J J, Sha W C. Immunity. 1999;11:423–432. doi: 10.1016/s1074-7613(00)80117-x. [DOI] [PubMed] [Google Scholar]
- 35.Richter G, Hayden-Ledbetter M, Irgang M, Ledbetter J A, Westermann J, Korner I, Daemen K, Clark E A, Aicher A, Pezzutto A. J Biol Chem. 2001;276:45686–45693. doi: 10.1074/jbc.M108509200. [DOI] [PubMed] [Google Scholar]
- 36.Lampugnani M G, Resnati M, Raiteri M, Pigott R, Pisacane A, Houen G, Ruco L P, Dejana E. J Cell Biol. 1992;118:1511–1522. doi: 10.1083/jcb.118.6.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy L L, Mazanet M M, Taylor A C, Mestas J, Hughes C C. Cell Immunol. 1999;194:150–161. doi: 10.1006/cimm.1999.1504. [DOI] [PubMed] [Google Scholar]
- 38.Sperling A I, Bluestone J A. Nat Immunol. 2001;2:573–574. doi: 10.1038/89709. [DOI] [PubMed] [Google Scholar]
- 39.Coyle A J, Lehar S, Lloyd C, Tian J, Delaney T, Manning S, Nguyen T, Burwell T, Schneider H, Gonzalo J A, et al. Immunity. 2000;13:95–105. doi: 10.1016/s1074-7613(00)00011-x. [DOI] [PubMed] [Google Scholar]
- 40.Lenschow D J, Walunas T L, Bluestone J A. Annu Rev Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]