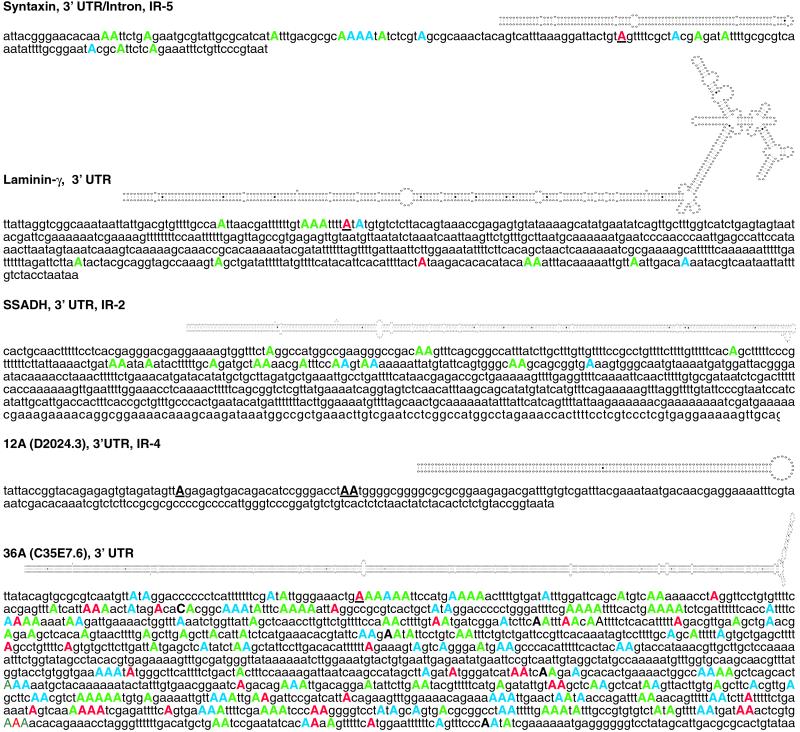
Figure 2.
Each of the newly identified C. elegans substrates is designated with a name, the location of editing within the mRNA, and the type of repetitive element, if one exists. Structures predicted by mfold are shown above the unedited sequence. Adenosines determined to be edited by cDNA sequencing are shown in uppercase, color-coded according to the percentage of the cDNA population that was edited at the site (red, >70%; blue, 40–70%; green, <40%); RNase T1 cleavage sites are underlined. Editing in syntaxin occurs within an alternatively spliced region that can be either an intron or a 3′ UTR. SSADH was found by virtue of its IR element, not by T1 cleavage, and only nucleotides 2538–2773 (complementary strand) of cosmid F45H10 were sequenced to determine editing sites. Editing sites were mapped by compiling sequence data from multiple PCR clones (SSADH), uncloned PCR populations (36A), or both types of data (syntaxin; laminin-γ); the editing sites of 12A have not been mapped, but three T1 cleavage sites were detected and are shown as black underlined capitals.