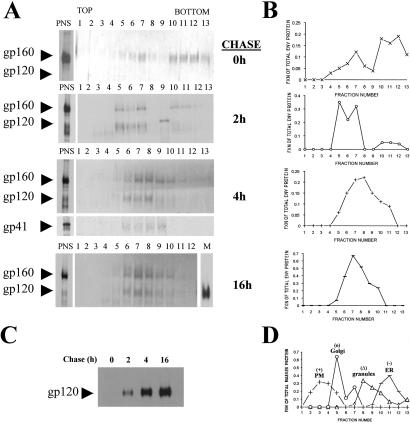
Figure 3.
HIV-1 Env traffics directly from Golgi to CTLA-4-containing granules. Lysates of radiolabeled H9-CTLA-4-GFP cells infected with vaccinia virus vPE16 (encoding HIV-1 Env) were resolved by density sucrose gradient centrifugation. (A) The images represent a 30-min radiolabeled pulse followed by 0-, 2-, 4-, or 16-h chase periods. An aliquot from the total postnuclear supernatant before resolution on the gradient is shown on the left side and immunoprecipitated for gp120 Env. Each fraction from the gradients was analyzed for the presence of radiolabeled gp120 Env by immunoprecipitation. In addition, the distribution of gp41 in the gradients at the 4-h chase is shown. (B) The autoradiographs from A were digitized and quantified with NIH IMAGE software. The fraction of total Env protein (gp120 + gp160) loaded onto the gradient and recovered from each gradient fraction is shown. (C) Immunoprecipitates of gp120 protein shed into the media from the pulse-chase experiments are shown on the autoradiograph. Equal volumes of culture supernatant were used from each time point. (D) The resolution of marker proteins of specific organelles on the gradient was analyzed by immunoblots. The resolved fractions were blotted for the marker proteins calreticulin (ER), MG160 (Golgi), CTLA-4 (regulated secretory granules), and CD4 (plasma membrane and endosomes). For each fraction, the amount of the marker is expressed as a percentage of total immunoreactivity across the gradient.