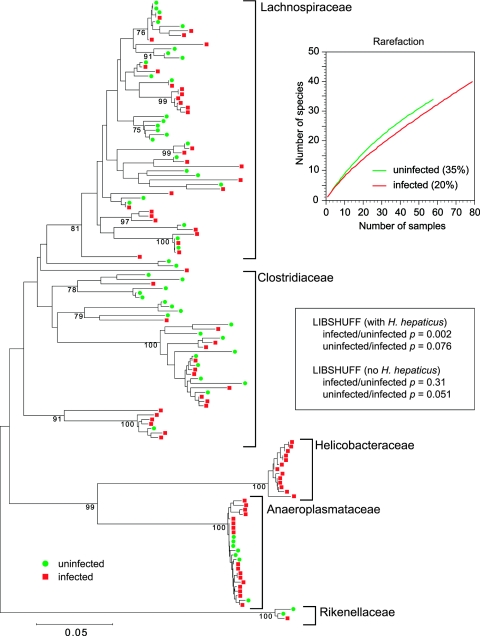
FIG.3.
Phylogenetic tree showing the distribution of 16S rRNA-encoding gene sequences from clone libraries constructed from cecal DNA samples obtained from an uninfected mouse (green circles) or a mouse 30 days after infection with H. hepaticus (red squares). Brackets outline major clusters of organisms. The scale bar represents evolutionary distance (5 substitutions per 100 nucleotides). The tree was constructed by neighbor-joining analysis using the MEGA program. Analysis was performed on a multiple-sequence alignment generated using the ARB suite of programs. Clones representing H. hepaticus were found only in the clone library constructed from DNA from the infected animal. The graph shows rarefaction curves for each library along with Good's coverage estimate in parentheses. The results of LIBSHUFF analysis (40) of the two libraries, with and without inclusion of H. hepaticus 16S rRNA-encoding gene sequences, are shown in the inset.