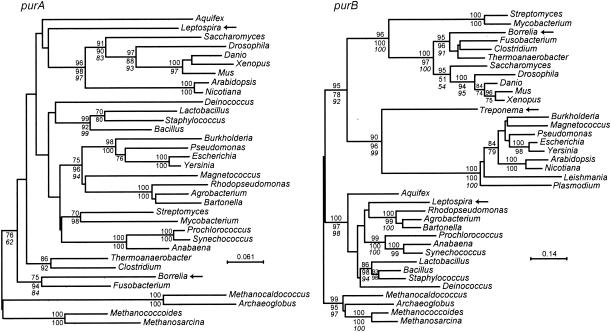
FIG. 2.
Phylograms from codon-based nucleotide sequence alignments with the third position excluded of the purA (left) and purB (right) genes of the following bacteria (B), archaea (A), and eukaryotes (E): Agrobacterium tumefaciens (B), Anabaena variabilis (B), Aquifex aeolicus (B), Arabidopsis thaliana (E), Archaeoglobus fulgidus (A), Bacillus subtilis (B), Bartonella henselae (B), Borrelia hermsii (B), Burkholderia pseudomallei (B), Clostridium perfringens (B), Danio rerio (E), Deinococcus radiodurans (B), Drosophila melanogaster (E), Escherichia coli K-12 (B), Fusobacterium nucleatum (B), Lactobacillus plantarum (B), Leishmania major (E), Leptospira interrogans serovar Copenhageni (B), Magnetococcus sp. strain MC-1 (B), Methanocaldococcus jannaschii (A), Methanococcoides burtonii (A), Methanosarcina acetivorans (A), Mus musculus (E), Mycobacterium tuberculosis H37Rv (B), Nicotiana tabacum (E), Plasmodium falciparum (E), Prochlorococcus marinus MIT9313 (B), Pseudomonas aeruginosa (B), Rhodopseudomonas palustris (B), Saccharomyces cerevisiae (E), Staphylococcus aureus N315 (B), Streptomyces coelicolor (B), Synechococcus sp. strain WH8102 (B), Thermoanaerobacter tengcongensis (B), Treponema denticola (B), Xenopus laevis (E), and Yersinia pestis CO92 (B). Arrows indicate spirochetes. Bootstrap values for nodes with ≥70% support by neighbor-joining distance (1,000 replicates; number above the line) and/or maximum-likelihood criteria (100 replicates; first number below the line) are shown. Italicized numbers below the line are bootstrap values (percent) by maximum parsimony criterion. Bars in each panel provide the scale for nucleotide distance.