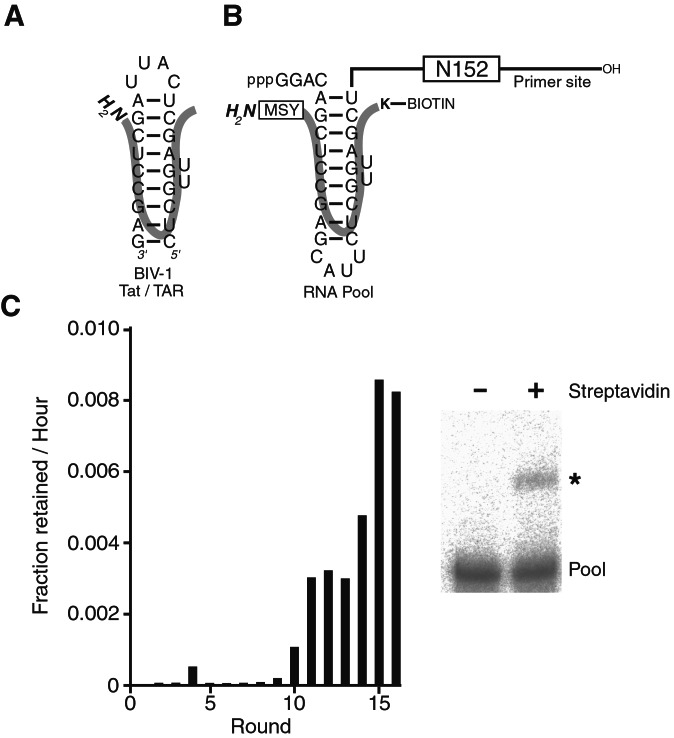
Figure 1.
(A) BIV-1 Tat/TAR. (B) Pool design. The block containing 152 random positions (N152) was flanked by defined segments used as primer-binding sites for reverse transcription and amplification of selected molecules. A circularly permuted BIV-1 TAR site was placed near the 5′-end of the RNA pool. The orientation of the BIV-1 Tat peptide bound to the TAR site is shown (thick line). A biotinylated lysine within the polypeptide served as an affinity tag for the selection of reacted molecules (K-Biotin). Residues were appended to the amino terminus of the wild-type peptide to increase the diversity of potential nucleophiles within the peptide (boxed MSY). (C) Progress of the selection. The amount of RNA retained on the streptavidin beads was quantitated during the course of selection. After 9 rounds, this amount increased to levels where the reacted product could be visualized on a denaturing polyacrylamide gel (asterisk).