Fig 1.
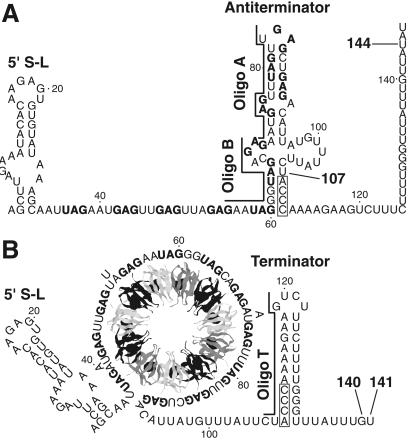
Model of the trpEDCFBA operon attenuation mechanism. During transcription, RNA polymerase pauses at U107. Pausing at this position is greatly stimulated by NusA. (A) In limiting tryptophan conditions, TRAP is not activated and does not bind to the nascent transcript. Eventually RNA polymerase overcomes the pause and resumes transcription. In this case, formation of the antiterminator prevents formation of the terminator, resulting in transcription readthrough into the trp structural genes. (B) In excess tryptophan conditions, TRAP is activated and binds to the (G/U)AG repeats soon after they are synthesized. NusA-stimulated pausing at U107 probably allows additional time for TRAP to bind. TRAP binding releases the paused RNA polymerase complex and transcription resumes. Bound TRAP prevents formation of the antiterminator, thereby allowing formation of the terminator, and hence the termination of transcription at G140 or U141. NusA-stimulated pausing also occurs at U144 in readthrough transcripts. This pausing event is probably involved in the trpE translation control mechanism. The 11 (G/U)AG repeats are indicated in boldface type. Numbering is from the start of transcription. The relative positions of oligonucleotide (oligo) A (nucleotides 70–84), oligo B (nucleotides 55–69), and oligo T (nucleotides 106–120) used in the pause hairpin competition studies are shown.