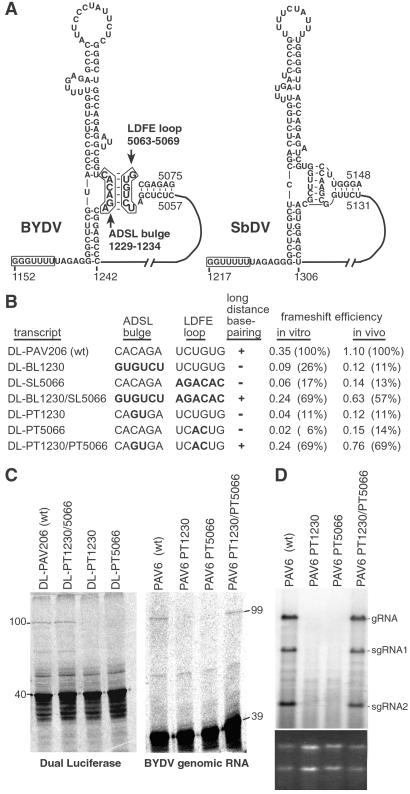
Fig 4.
Frameshifting and replication of BYDV mutants that destroy and restore the long-distance base pairing. (A) Predicted secondary structures (MFOLD) of BYDV and the related SbDV RNAs showing the base-pairing interaction (boxed in BYDV structure) of the long-distance frameshift element (LDFE) with the bulge in the adjacent downstream stem loop (ADSL bulge). (B) Frameshift efficiencies of dual luciferase constructs with mutations in ADSL bulge and/or LDFE that disrupt and restore potential base pairing. Frameshift efficiencies are averages from three independent experiments. The nucleotide sequence for the two regions for each mutant is shown with altered bases in bold. (C) PAGE of wheat germ translation products of BYDV dual luciferase (100-kDa frameshift product) and of full-length viral (PAV6) constructs containing the same sets of two-base mutations in the ADSL and/or LDFE. (D) Northern blot hybridization of RNA from oat protoplasts inoculated with the same wild-type and mutant PAV6 transcripts used in C. BYDV genomic (gRNA) and subgenomic RNAs (sgRNA) produced during virus replication are marked. (Lower) Ethidium bromide staining of gel used for the above blot as a control for gel loading. This experiment was repeated three times with the same results.