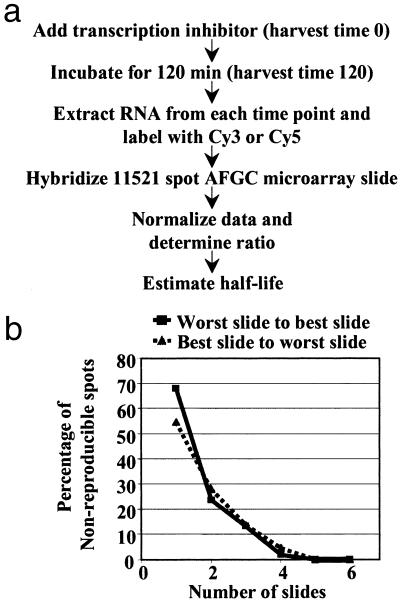
Fig 1.
Strategy for monitoring mRNA stability using cDNA microarrays. (a) RNA samples corresponding to 0 and 120 min after the addition of the transcriptional inhibitor cordycepin were labeled with Cy3 and Cy5, respectively, and used to hybridize 11K microarray slides. Each pair of RNA samples was reverse labeled for a separate microarray hybridization. These hybridizations were performed with samples from three independent cordycepin treatments for a final data set of six slides. Half-life values were then estimated from the normalized ratios. (b) Nonreproducible spots decrease as a function of the number of slides, nearly leveling out when the data from four slides is combined. The quality of the slides, best to worst or worst to best based on the extent of visible gradients (see Materials and Methods), does not significantly affect the reproducibility of the data when two or more slides are considered, although the curves are slightly steeper with better slides.