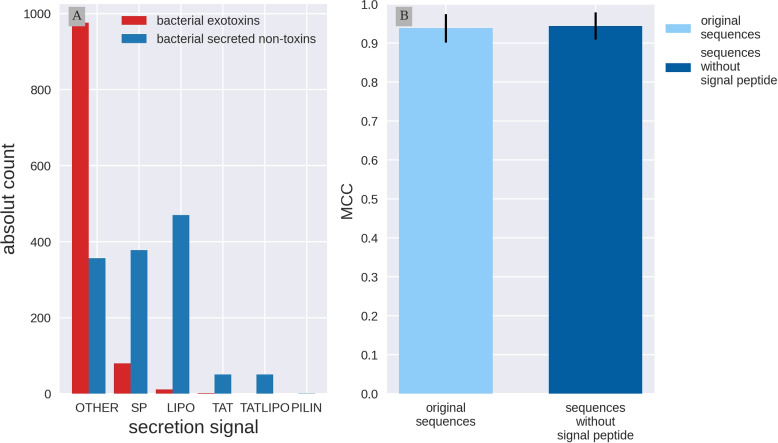
Fig. 3.
Signal Peptides do not influence toxin prediction Panel A: Exploration of signal peptides predicted with SignalP-6.0. Bacterial exotoxins are in red, the bacterial secreted non-toxins are in blue. Signal Peptides distinguish between two translocation routs Sec and Tat and three Signal Peptidases SPI-III. Prediction include SP: Sec/SPI, LIPO: Sec/SPII, TAT: Tat/SPI, LIPOTAT: Tat/SPII, PILIN: Sec/SPIII and OTHER indicates no known signal peptides. The majority of exotoxins does not have a predicted signal peptide. Panel B: Performance comparison with and without signal Peptides. Model architecture was introduced in Fig. 2 Support Vector Classifier (SVC) using the first 20 Principal Components calculated on per protein protT5 embeddings (Embs20). Two versions of the test set are compared. Light blue are the original test set sequences. Dark blue: the test sequences without the predicted signal peptides. Embs20/SVC performs equally well on both test set versions