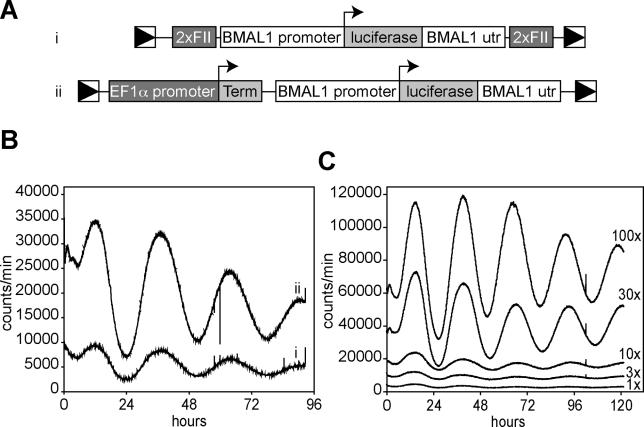
Figure 1. Circadian Bioluminescence Can Be Recorded in Fibroblasts Infected with a Lentiviral Luciferase Expression Vector.
(A) Circadian reporter constructs used in these studies. Each contains the mouse Bmal1 promoter, the firefly luciferase coding region, and the Bmal1 3′UTR, flanked by the long terminal repeats (LTRs) of a lentiviral packaging vector. In (i), a dimerized chick β-globin FII element is inserted between each LTR and adjacent Bmal1 sequences. In (ii), a DNA segment composed of the EF1α promoter and a SV40 terminator is inserted between the upstream LTR and Bmal1 promoter, and the gfp coding region between the Bmal1 UTR and the downstream viral LTR.
(B) 3T3 cells were infected with the lentiviral vectors shown above, and equivalent infection levels were verified by real-time PCR to detect integrated viruses. Four days after infection, cells were shocked with dexamethasone to synchronize circadian rhythms, and luciferase output was measured by real-time luminometry.
(C) 3T3 cells were infected with different concentrations of the lentiviral reporter vector ii (see [A]), and circadian rhythms were measured as in (B). 10× represents unconcentrated filtered viral supernatant, 1× represents a 10× dilution of this, and 100× was a 10× concentration by ultracentrifugation. The number of viral infection units/plate were approximately 10,000 (1×), 30,000 (3×), 100,000 (10×), 300,000 (30×), and 1,000,000 (100×).