Abstract
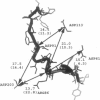
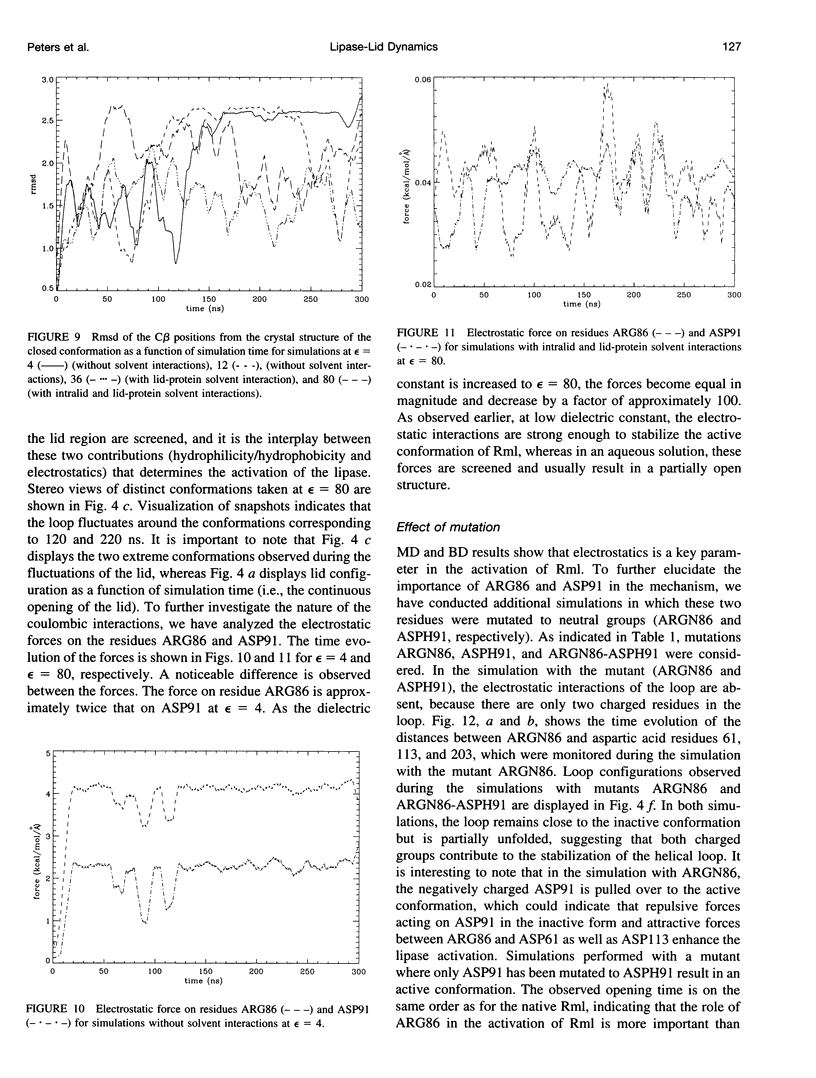
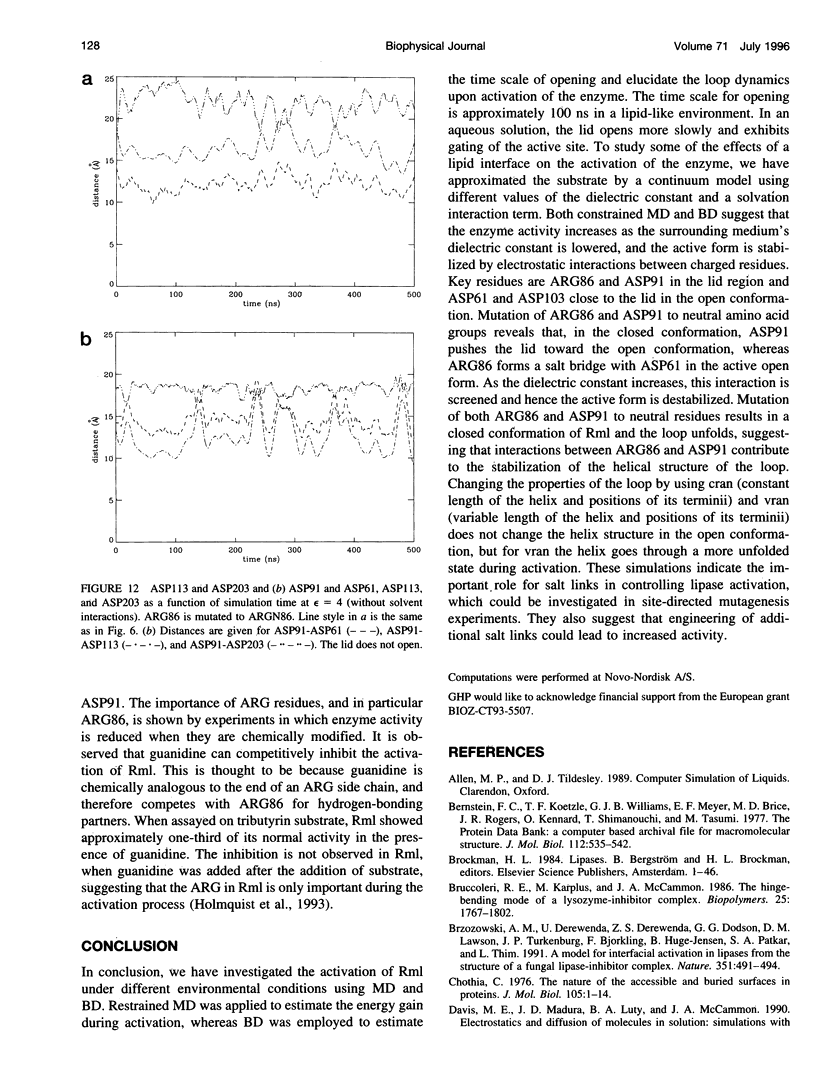
Interfacial activation of Rhizomucor miehei lipase is accompanied by a hinge-type motion of a single helix (residues 83-94) that acts as a lid over the active site. Activation of the enzyme involves the displacement of the lid to expose the active site, suggesting that the dynamics of the lid could be of mechanistic and kinetic importance. To investigate possible activation pathways and to elucidate the effect of a hydrophobic environment (as would be provided by a lipid membrane) on the lid opening, we have applied molecular dynamics and Brownian dynamics techniques. Our results indicate that the lipase activation is enhanced in a hydrophobic environment. In nonpolar low-dielectric surroundings, the lid opens in approximately 100 ns in the BD simulations. In polar high-dielectric (aqueous) surroundings, the lid does not always open up in simulations of up to 900 ns duration, but it does exhibit some gating motion, suggesting that the enzyme molecule may exist in a partially active form before the catalytic reaction. The activation is controlled by the charged residues ARG86 and ASP91. In the inactive conformation, ASP91 experiences repulsive forces and pushes the lid toward the open conformation. Upon activation ARG86 approaches ASP61, and in the active conformation, these residues form a salt bridge that stabilizes the open conformation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bruccoleri R. E., Karplus M., McCammon J. A. The hinge-bending mode of a lysozyme-inhibitor complex. Biopolymers. 1986 Sep;25(9):1767–1802. doi: 10.1002/bip.360250916. [DOI] [PubMed] [Google Scholar]
- Brzozowski A. M., Derewenda U., Derewenda Z. S., Dodson G. G., Lawson D. M., Turkenburg J. P., Bjorkling F., Huge-Jensen B., Patkar S. A., Thim L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991 Jun 6;351(6326):491–494. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976 Jul 25;105(1):1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Derewenda U., Brzozowski A. M., Lawson D. M., Derewenda Z. S. Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase. Biochemistry. 1992 Feb 11;31(5):1532–1541. doi: 10.1021/bi00120a034. [DOI] [PubMed] [Google Scholar]
- Derewenda U., Swenson L., Wei Y., Green R., Kobos P. M., Joerger R., Haas M. J., Derewenda Z. S. Conformational lability of lipases observed in the absence of an oil-water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J Lipid Res. 1994 Mar;35(3):524–534. [PubMed] [Google Scholar]
- Derewenda Z. S. A twist in the tale of lipolytic enzymes. Nat Struct Biol. 1995 May;2(5):347–349. doi: 10.1038/nsb0595-347. [DOI] [PubMed] [Google Scholar]
- Derewenda Z. S., Derewenda U., Dodson G. G. The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 A resolution. J Mol Biol. 1992 Oct 5;227(3):818–839. doi: 10.1016/0022-2836(92)90225-9. [DOI] [PubMed] [Google Scholar]
- Falzone C. J., Wright P. E., Benkovic S. J. Dynamics of a flexible loop in dihydrofolate reductase from Escherichia coli and its implication for catalysis. Biochemistry. 1994 Jan 18;33(2):439–442. doi: 10.1021/bi00168a007. [DOI] [PubMed] [Google Scholar]
- Holmquist M., Norin M., Hult K. The role of arginines in stabilizing the active open-lid conformation of Rhizomucor miehei lipase. Lipids. 1993 Aug;28(8):721–726. doi: 10.1007/BF02535993. [DOI] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature. 1979 Feb 8;277(5696):491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Kempner E. S. Movable lobes and flexible loops in proteins. Structural deformations that control biochemical activity. FEBS Lett. 1993 Jul 12;326(1-3):4–10. doi: 10.1016/0014-5793(93)81749-p. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976 Jun 14;104(1):59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Levitt M., Warshel A. Computer simulation of protein folding. Nature. 1975 Feb 27;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M., Wolynes P. G. The hinge-bending mode in lysozyme. Nature. 1976 Jul 22;262(5566):325–326. doi: 10.1038/262325a0. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Northrup S. H. Gated binding of ligands to proteins. Nature. 1981 Sep 24;293(5830):316–317. doi: 10.1038/293316a0. [DOI] [PubMed] [Google Scholar]
- Muderhwa J. M., Brockman H. L. Lateral lipid distribution is a major regulator of lipase activity. Implications for lipid-mediated signal transduction. J Biol Chem. 1992 Dec 5;267(34):24184–24192. [PubMed] [Google Scholar]
- Norin M., Haeffner F., Hult K., Edholm O. Molecular dynamics simulations of an enzyme surrounded by vacuum, water, or a hydrophobic solvent. Biophys J. 1994 Aug;67(2):548–559. doi: 10.1016/S0006-3495(94)80515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norin M., Olsen O., Svendsen A., Edholm O., Hult K. Theoretical studies of Rhizomucor miehei lipase activation. Protein Eng. 1993 Nov;6(8):855–863. doi: 10.1093/protein/6.8.855. [DOI] [PubMed] [Google Scholar]
- Peters G. H., Toxvaerd S., Larsen N. B., Bjørnholm T., Schaumburg K., Kjaer K. Structure and dynamics of lipid monolayers: implications for enzyme catalysed lipolysis. Nat Struct Biol. 1995 May;2(5):395–401. doi: 10.1038/nsb0595-395. [DOI] [PubMed] [Google Scholar]
- Philippopoulos M., Xiang Y., Lim C. Identifying the mechanism of protein loop closure: a molecular dynamics simulation of the Bacillus stearothermophilus LDH loop in solution. Protein Eng. 1995 Jun;8(6):565–573. doi: 10.1093/protein/8.6.565. [DOI] [PubMed] [Google Scholar]
- Piéroni G., Gargouri Y., Sarda L., Verger R. Interactions of lipases with lipid monolayers. Facts and questions. Adv Colloid Interface Sci. 1990 Sep;32(4):341–378. doi: 10.1016/0001-8686(90)80023-s. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Richards F. M. Packing of alpha-helices: geometrical constraints and contact areas. J Mol Biol. 1978 Mar 15;119(4):537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- Thuren T. A model for the molecular mechanism of interfacial activation of phospholipase A2 supporting the substrate theory. FEBS Lett. 1988 Feb 29;229(1):95–99. doi: 10.1016/0014-5793(88)80805-6. [DOI] [PubMed] [Google Scholar]
- Wade R. C., Davis M. E., Luty B. A., Madura J. D., McCammon J. A. Gating of the active site of triose phosphate isomerase: Brownian dynamics simulations of flexible peptide loops in the enzyme. Biophys J. 1993 Jan;64(1):9–15. doi: 10.1016/S0006-3495(93)81335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Davenport R. C. Comparison of the refined crystal structures of liganded and unliganded chicken, yeast and trypanosomal triosephosphate isomerase. J Mol Biol. 1992 Apr 20;224(4):1115–1126. doi: 10.1016/0022-2836(92)90473-w. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Vriend G., Nauche S., Hol W. G. Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex. J Mol Biol. 1991 Aug 20;220(4):995–1015. doi: 10.1016/0022-2836(91)90368-g. [DOI] [PubMed] [Google Scholar]
- Williams J. C., McDermott A. E. Dynamics of the flexible loop of triosephosphate isomerase: the loop motion is not ligand gated. Biochemistry. 1995 Jul 4;34(26):8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]