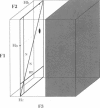
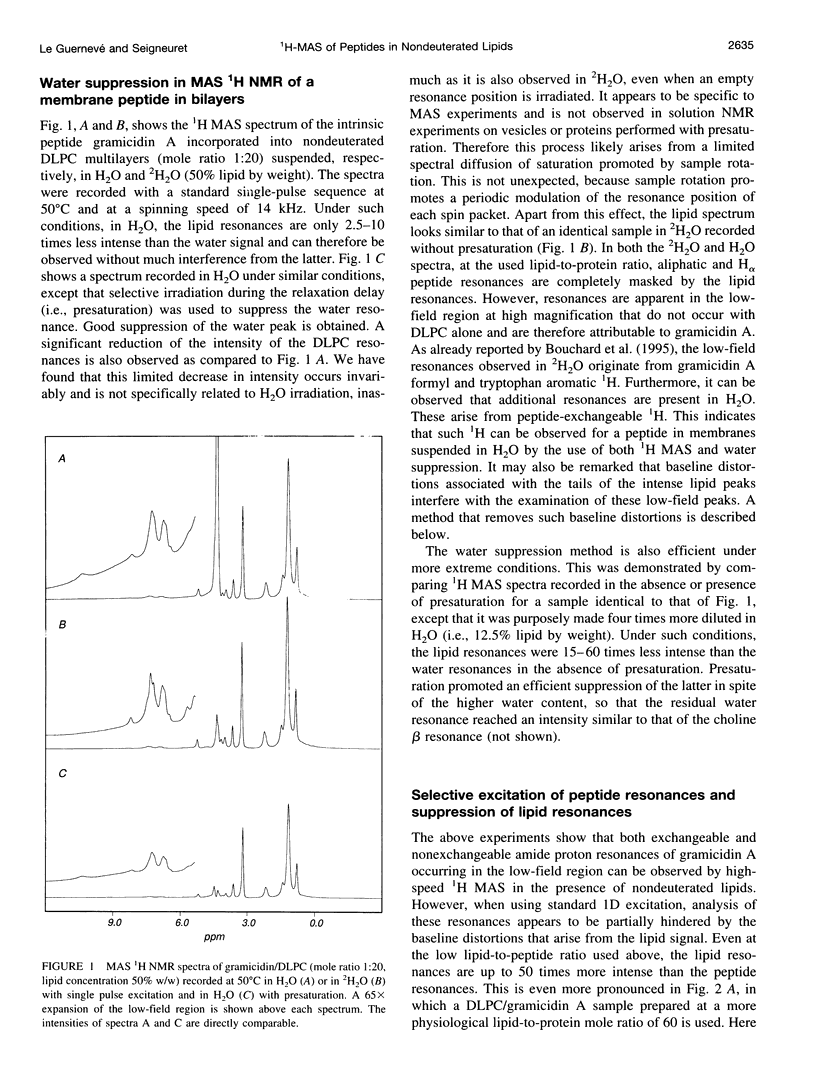
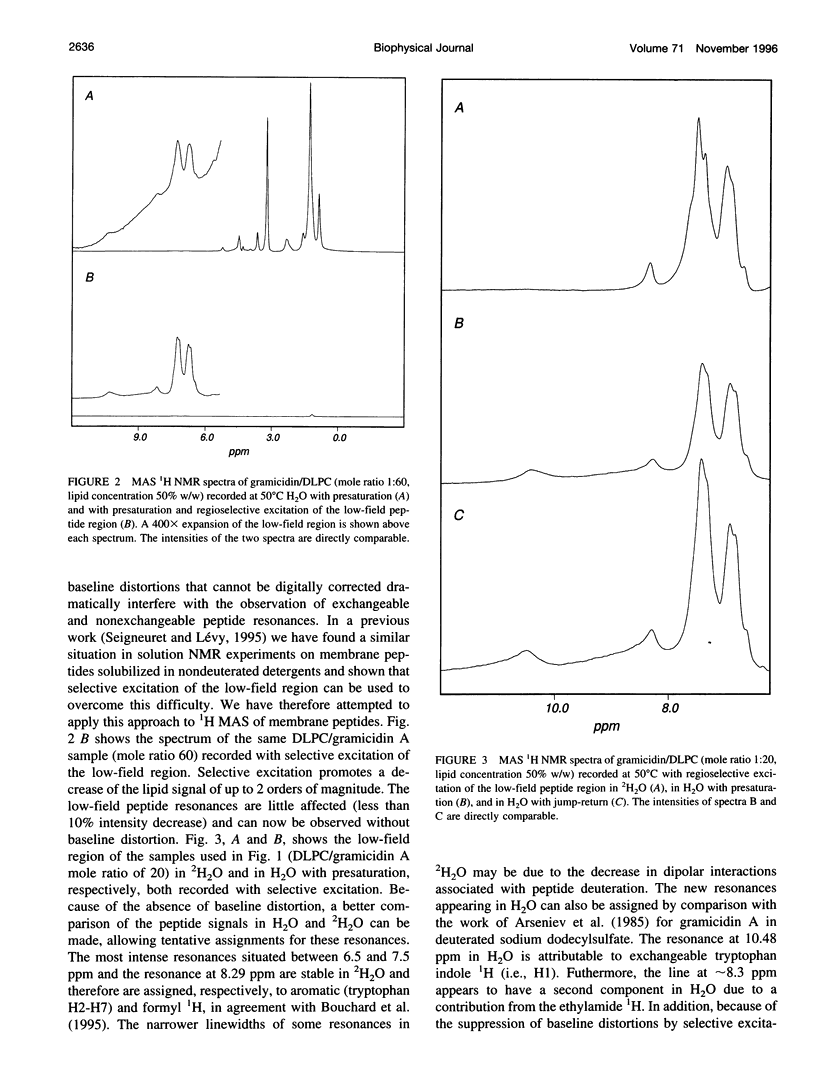
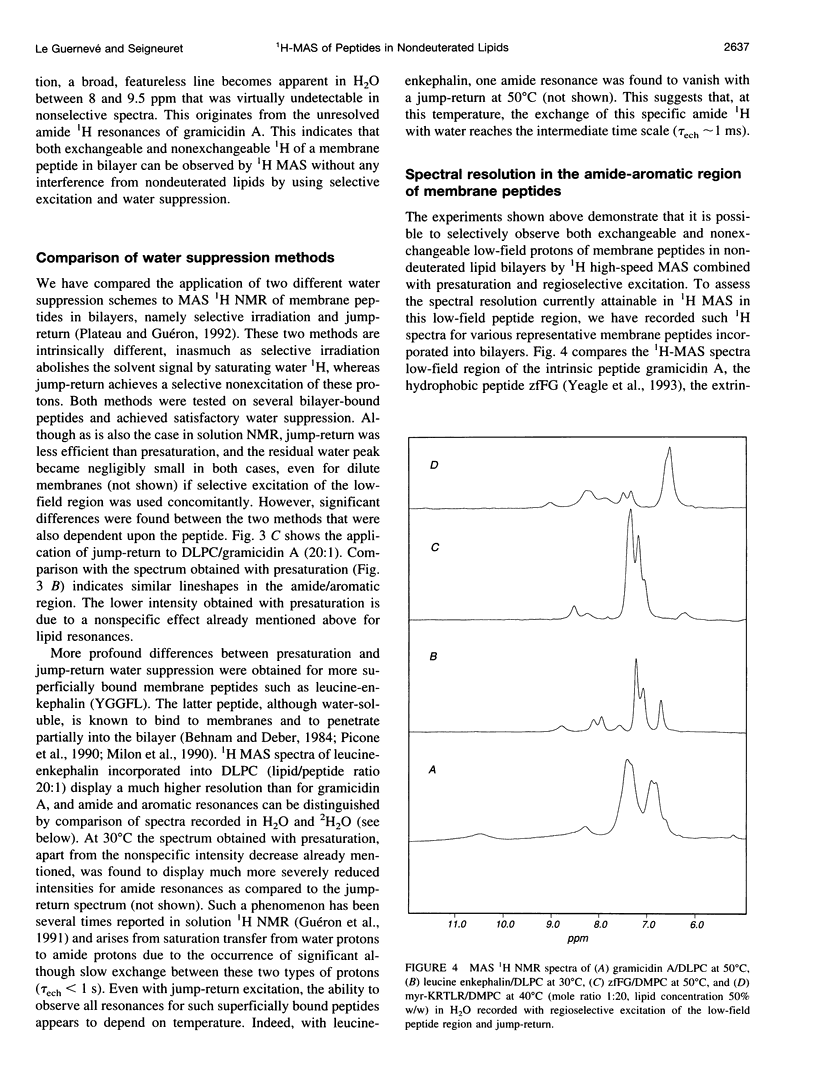
Abstract
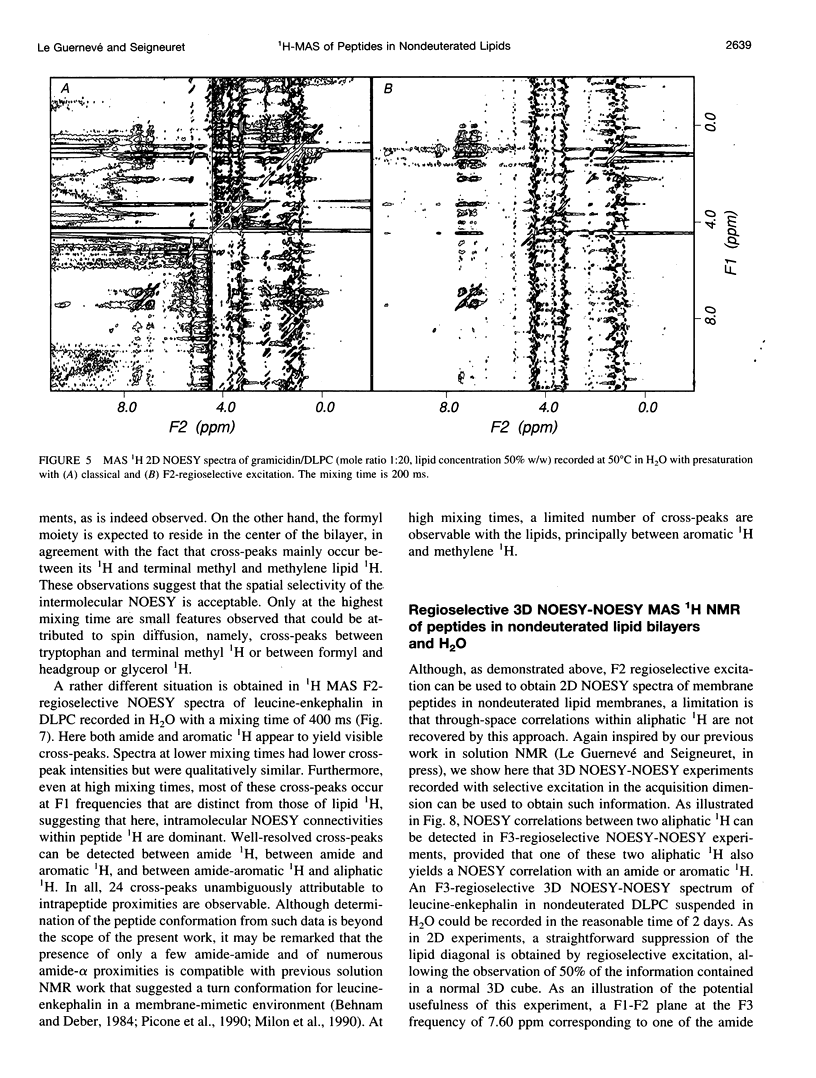
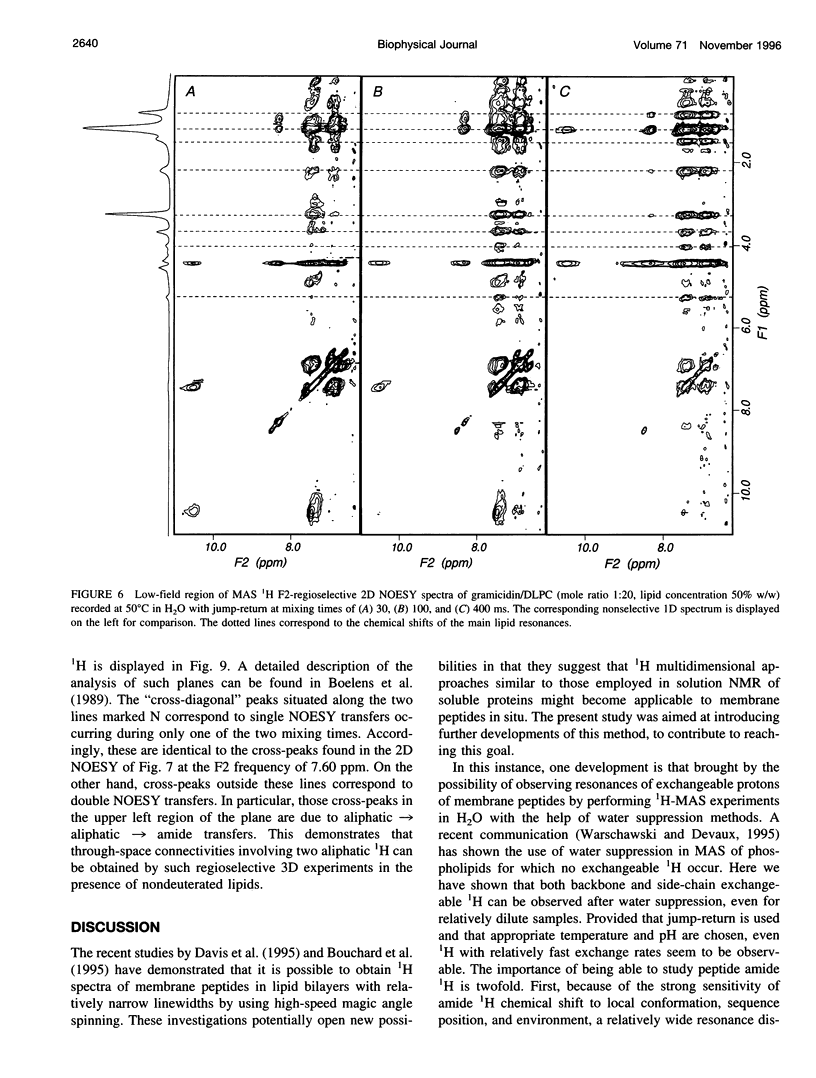
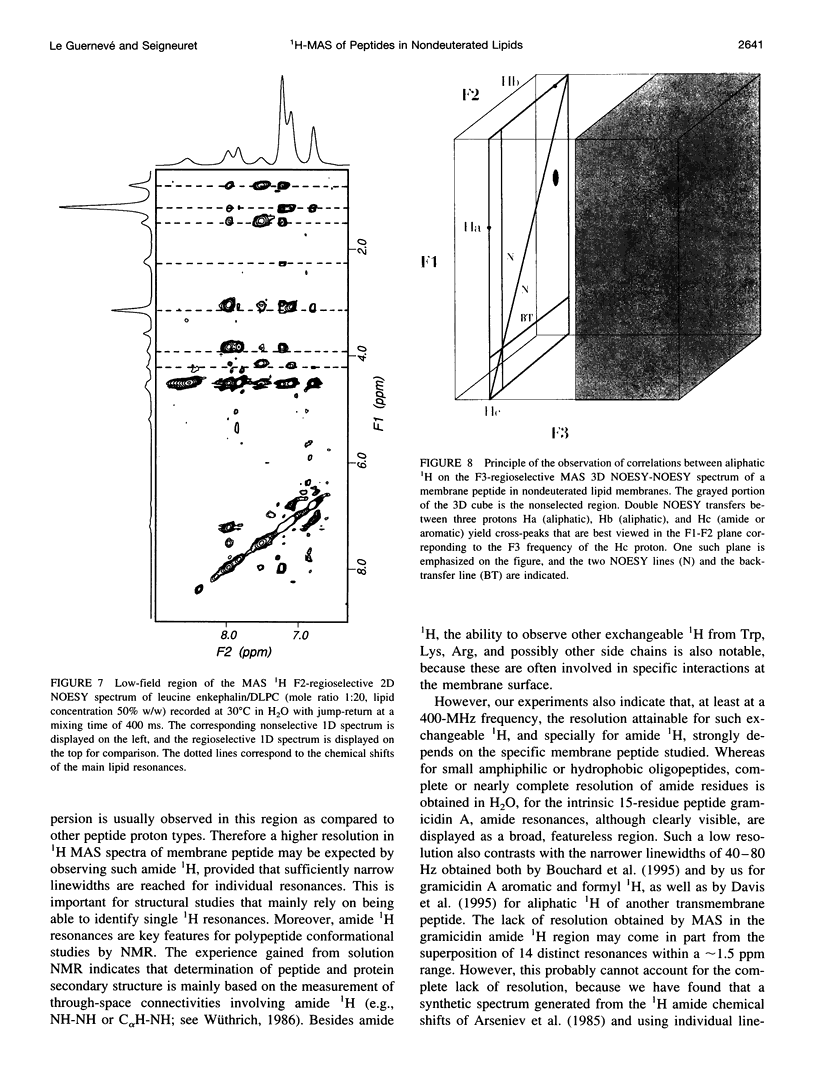
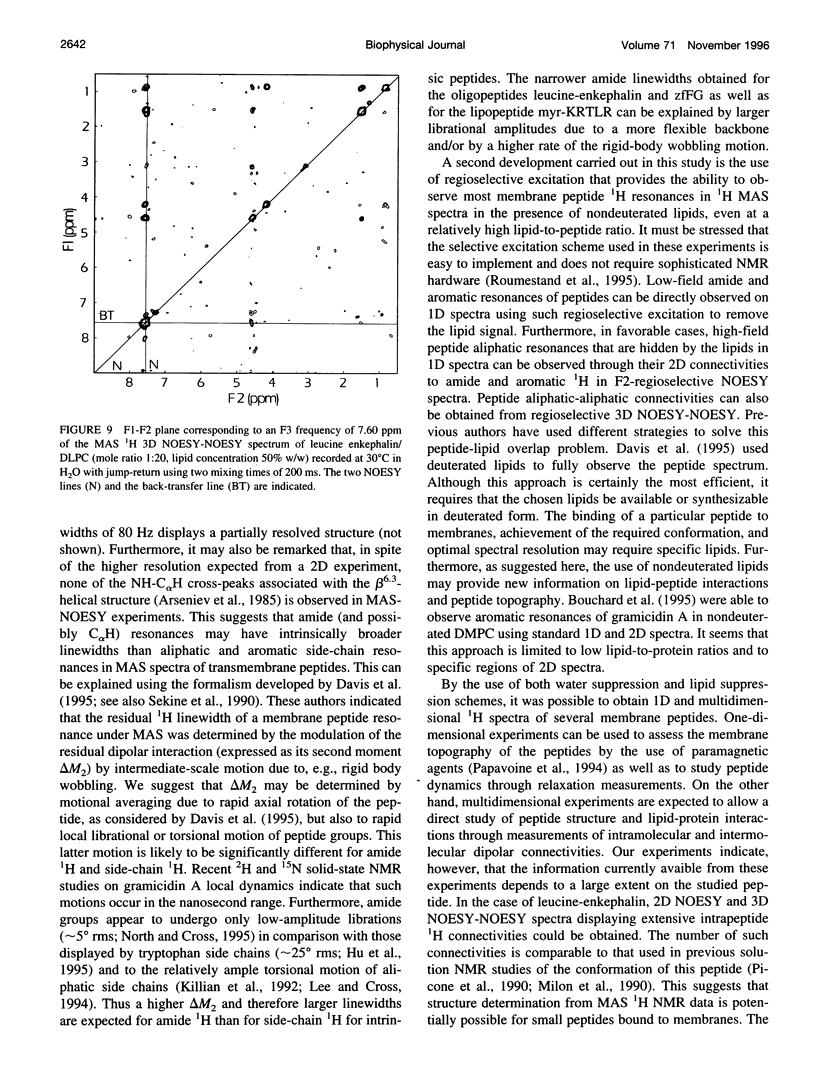
High-speed (14 kHz) solid-state magic angle spinning (MAS) 1H NMR has been applied to several membrane peptides incorporated into nondeuterated dilauroyl or dimyristoylphosphatidylcholine membranes suspended in H2O. It is shown that solvent suppression methods derived from solution NMR, such as presaturation or jump-return, can be used to reduce water resonance, even at relatively high water content. In addition, regioselective excitation of 1H peptide resonances promotes an efficient suppression of lipid resonances, even in cases where these are initially two orders of magnitude more intense. As a consequence, 1H MAS spectra of the peptide low-field region are obtained without interference from water and lipid signals. These display resonances from amide and other exchangeable 1H as well as from aromatic nonexchangeable 1H. The spectral resolution depends on the specific types of resonance and membrane peptide. For small amphiphilic or hydrophobic oligopeptides, resolution of most individual amide resonance is achieved, whereas for the transmembrane peptide gramicidin A, an unresolved amide spectrum is obtained. Partial resolution of aromatic 1H occurs in all cases. Multidimensional 1H-MAS spectra of membrane peptides can also be obtained by using water suppression and regioselective excitation. For gramicidin A, F2-regioselective 2D nuclear Overhauser effect spectroscopy (NOESY) spectra are dominated by intermolecular through-space connectivities between peptide aromatic or formyl 1H and lipid 1H. These appear to be compatible with the known structure and topography of the gramicidin pore. On the other hand, for the amphiphilic peptide leucine-enkephalin, F2-regioselective NOESY spectra mostly display cross-peaks originating from though-space proximities of amide or aromatic 1H with themselves and with aliphatic 1H. F3-regioselective 3D NOESY-NOESY spectra can be used to obtain through-space correlations within aliphatic 1H. Such intrapeptide proximities should allow determination of the conformation of the peptide in membranes. It is suggested that high-speed MAS multidimensional 1H NMR of peptides in nondeuterated membranes and in H2O can be used for studies of both peptide structure and lipid-peptide interactions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arseniev A. S., Barsukov I. L., Bystrov V. F., Lomize A. L., Ovchinnikov YuA 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single-stranded helices. FEBS Lett. 1985 Jul 8;186(2):168–174. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- Bechinger B., Zasloff M., Opella S. J. Structure and orientation of the antibiotic peptide magainin in membranes by solid-state nuclear magnetic resonance spectroscopy. Protein Sci. 1993 Dec;2(12):2077–2084. doi: 10.1002/pro.5560021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnam B. A., Deber C. M. Evidence for a folded conformation of methionine- and leucine-enkephalin in a membrane environment. J Biol Chem. 1984 Dec 10;259(23):14935–14940. [PubMed] [Google Scholar]
- Bouchard M., Davis J. H., Auger M. High-speed magic angle spinning solid-state 1H nuclear magnetic resonance study of the conformation of gramicidin A in lipid bilayers. Biophys J. 1995 Nov;69(5):1933–1938. doi: 10.1016/S0006-3495(95)80063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Van Gorkom L. C., Epand R. M., Stark R. E. Nuclear magnetic resonance studies of lipid hydration in monomethyldioleoylphosphatidylethanolamine dispersions. Biophys J. 1996 Mar;70(3):1412–1418. doi: 10.1016/S0006-3495(96)79700-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzet F., McDermott A., Gebhard R., van der Hoef K., Spijker-Assink M. B., Herzfeld J., Lugtenburg J., Levitt M. H., Griffin R. G. Determination of membrane protein structure by rotational resonance NMR: bacteriorhodopsin. Science. 1991 Feb 15;251(4995):783–786. doi: 10.1126/science.1990439. [DOI] [PubMed] [Google Scholar]
- Davis J. H., Auger M., Hodges R. S. High resolution 1H nuclear magnetic resonance of a transmembrane peptide. Biophys J. 1995 Nov;69(5):1917–1932. doi: 10.1016/S0006-3495(95)80062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995 Nov;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Henry G. D., Sykes B. D. Methods to study membrane protein structure in solution. Methods Enzymol. 1994;239:515–535. doi: 10.1016/s0076-6879(94)39020-7. [DOI] [PubMed] [Google Scholar]
- Hu W., Lazo N. D., Cross T. A. Tryptophan dynamics and structural refinement in a lipid bilayer environment: solid state NMR of the gramicidin channel. Biochemistry. 1995 Oct 31;34(43):14138–14146. doi: 10.1021/bi00043a019. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Killian J. A., Prasad K. U., Hains D., Urry D. W. The membrane as an environment of minimal interconversion. A circular dichroism study on the solvent dependence of the conformational behavior of gramicidin in diacylphosphatidylcholine model membranes. Biochemistry. 1988 Jun 28;27(13):4848–4855. doi: 10.1021/bi00413a040. [DOI] [PubMed] [Google Scholar]
- Killian J. A., Taylor M. J., Koeppe R. E., 2nd Orientation of the valine-1 side chain of the gramicidin transmembrane channel and implications for channel functioning. A 2H NMR study. Biochemistry. 1992 Nov 24;31(46):11283–11290. doi: 10.1021/bi00161a004. [DOI] [PubMed] [Google Scholar]
- Lee K. C., Cross T. A. Side-chain structure and dynamics at the lipid-protein interface: Val1 of the gramicidin A channel. Biophys J. 1994 May;66(5):1380–1387. doi: 10.1016/S0006-3495(94)80928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon A., Miyazawa T., Higashijima T. Transferred nuclear Overhauser effect analyses of membrane-bound enkephalin analogues by 1H nuclear magnetic resonance: correlation between activities and membrane-bound conformations. Biochemistry. 1990 Jan 9;29(1):65–75. doi: 10.1021/bi00453a009. [DOI] [PubMed] [Google Scholar]
- Mosior M., McLaughlin S. Peptides that mimic the pseudosubstrate region of protein kinase C bind to acidic lipids in membranes. Biophys J. 1991 Jul;60(1):149–159. doi: 10.1016/S0006-3495(91)82038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North C. L., Barranger-Mathys M., Cafiso D. S. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys J. 1995 Dec;69(6):2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North C. L., Cross T. A. Correlations between function and dynamics: time scale coincidence for ion translocation and molecular dynamics in the gramicidin channel backbone. Biochemistry. 1995 May 2;34(17):5883–5895. doi: 10.1021/bi00017a018. [DOI] [PubMed] [Google Scholar]
- Oldfield E., Bowers J. L., Forbes J. High-resolution proton and carbon-13 NMR of membranes: why sonicate? Biochemistry. 1987 Nov 3;26(22):6919–6923. doi: 10.1021/bi00396a009. [DOI] [PubMed] [Google Scholar]
- Opella S. J., Kim Y., McDonnell P. Experimental nuclear magnetic resonance studies of membrane proteins. Methods Enzymol. 1994;239:536–560. doi: 10.1016/s0076-6879(94)39021-5. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Wüthrich K. Protein hydration in aqueous solution. Science. 1991 Nov 15;254(5034):974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- Papavoine C. H., Konings R. N., Hilbers C. W., van de Ven F. J. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 1994 Nov 8;33(44):12990–12997. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- Picone D., D'Ursi A., Motta A., Tancredi T., Temussi P. A. Conformational preferences of [Leu5]enkephalin in biomimetic media. Investigation by 1H NMR. Eur J Biochem. 1990 Sep 11;192(2):433–439. doi: 10.1111/j.1432-1033.1990.tb19245.x. [DOI] [PubMed] [Google Scholar]
- Prosser R. S., Daleman S. I., Davis J. H. The structure of an integral membrane peptide: a deuterium NMR study of gramicidin. Biophys J. 1994 May;66(5):1415–1428. doi: 10.1016/S0006-3495(94)80932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser R. S., Davis J. H. Dynamics of an integral membrane peptide: a deuterium NMR relaxation study of gramicidin. Biophys J. 1994 May;66(5):1429–1440. doi: 10.1016/S0006-3495(94)80933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. R., 2nd, Landis G. C. Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry. 1995 Mar 28;34(12):4030–4040. doi: 10.1021/bi00012a022. [DOI] [PubMed] [Google Scholar]
- Seigneuret M., Lévy D. A high-resolution 1H NMR approach for structure determination of membrane peptides and proteins in non-deuterated detergent: application to mastoparan X solubilized in n-octylglucoside. J Biomol NMR. 1995 Jun;5(4):345–352. doi: 10.1007/BF00182276. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Bormann B. J. Determination of helix-helix interactions in membranes by rotational resonance NMR. Proc Natl Acad Sci U S A. 1995 Jan 17;92(2):488–491. doi: 10.1073/pnas.92.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O., Hamilton J., Salmon A., Bormann B. J. Rotational resonance NMR determination of intra- and intermolecular distance constraints in dipalmitoylphosphatidylcholine bilayers. Biochemistry. 1994 May 24;33(20):6327–6333. doi: 10.1021/bi00186a036. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Peersen O. B. Solid-state NMR approaches for studying membrane protein structure. Annu Rev Biophys Biomol Struct. 1992;21:25–47. doi: 10.1146/annurev.bb.21.060192.000325. [DOI] [PubMed] [Google Scholar]
- Volke F., Pampel A. Membrane hydration and structure on a subnanometer scale as seen by high resolution solid state nuclear magnetic resonance: POPC and POPC/C12EO4 model membranes. Biophys J. 1995 May;68(5):1960–1965. doi: 10.1016/S0006-3495(95)80373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warschawski D. E., Devaux P. F. Water suppression in 1H MASS NMR spectra of lipids and biological membranes. J Magn Reson B. 1995 Jan;106(1):76–79. doi: 10.1006/jmrb.1995.1012. [DOI] [PubMed] [Google Scholar]
- Woolf T. B., Roux B. Structure, energetics, and dynamics of lipid-protein interactions: A molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996 Jan;24(1):92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Yeagle P. L., Dentino A. R., Smith F. T., Spooner P., Watts A. The antiviral peptide carbobenzoxy-D-phenylalanyl-L-phenylalanylglycine changes the average conformation of phospholipids in membranes. Biochemistry. 1993 Nov 16;32(45):12197–12202. doi: 10.1021/bi00096a032. [DOI] [PubMed] [Google Scholar]