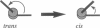Abstract
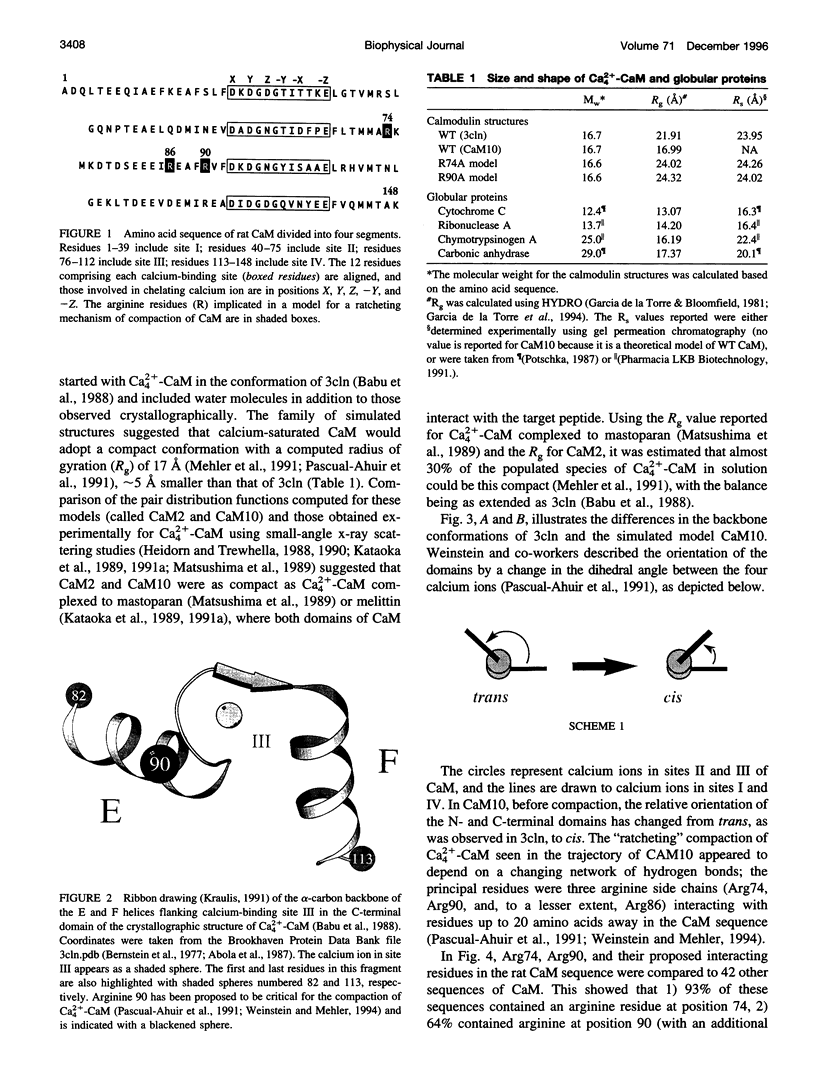
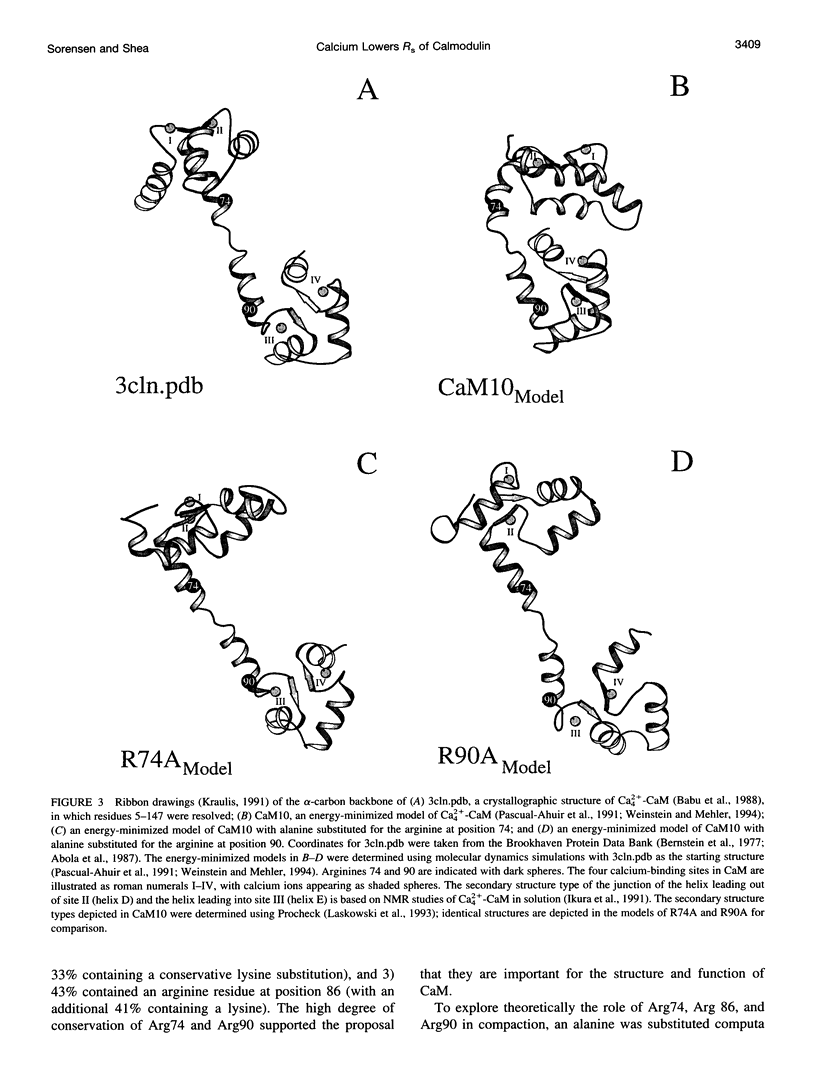
Calmodulin (CaM) is an intracellular cooperative calcium-binding protein essential for activating many diverse target proteins. Biophysical studies of the calcium-induced conformational changes of CaM disagree on the structure of the linker between domains and possible orientations of the domains. Molecular dynamics studies have predicted that Ca4(2+)CaM is in equilibrium between an extended and compact conformation and that Arg74 and Arg90 are critical to the compaction process. In this study gel permeation chromatography was used to resolve calcium-induced changes in the hydrated shape of CaM at pH 7.4 and 5.6. Results showed that mutation of Arg 74 to Ala increases the R(s) as predicted; however, the average separation of domains in Ca4(2+)-CaM was larger than predicted by molecular dynamics. Mutation of Arg90 to Ala or Gly affected the dimensions of apo-CaM more than those of Ca4(2+)-CaM. Calcium binding to CaM and mutants (R74A-CaM, R90A-CaM, and R90G-CaM) lowered the Stokes radius (R(s)). Differences between R(s) values reported here and Rg values determined by small-angle x-ray scattering studies illustrate the importance of using multiple techniques to explore the solution properties of a flexible protein such as CaM.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ACKERS G. K. MOLECULAR EXCLUSION AND RESTRICTED DIFFUSION PROCESSES IN MOLECULAR-SIEVE CHROMATOGRAPHY. Biochemistry. 1964 May;3:723–730. doi: 10.1021/bi00893a021. [DOI] [PubMed] [Google Scholar]
- ACKERS G. K., THOMPSON T. E. DETERMINATION OF STOICHIOMETRY AND EQUILIBRIUM CONSTANTS FOR REVERSIBLY ASSOCIATING SYSTEMS BY MOLECULAR SIEVE CHROMATOGRAPHY. Proc Natl Acad Sci U S A. 1965 Feb;53:342–349. doi: 10.1073/pnas.53.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackers G. K. Analytical gel chromatography of proteins. Adv Protein Chem. 1970;24:343–446. doi: 10.1016/s0065-3233(08)60245-4. [DOI] [PubMed] [Google Scholar]
- Andrews P. Estimation of molecular size and molecular weights of biological compounds by gel filtration. Methods Biochem Anal. 1970;18:1–53. [PubMed] [Google Scholar]
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965 Sep;96(3):595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulabaugh A., Niemczura W. P., Gibbons W. A. High field proton NMR studies of tryptic fragments of calmodulin: a comparison with the native protein. Biochem Biophys Res Commun. 1984 Jan 13;118(1):225–232. doi: 10.1016/0006-291x(84)91090-8. [DOI] [PubMed] [Google Scholar]
- Babu Y. S., Bugg C. E., Cook W. J. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988 Nov 5;204(1):191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992 Jun 16;31(23):5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- Bayley P. M., Martin S. R. The alpha-helical content of calmodulin is increased by solution conditions favouring protein crystallisation. Biochim Biophys Acta. 1992 Nov 10;1160(1):16–21. doi: 10.1016/0167-4838(92)90034-b. [DOI] [PubMed] [Google Scholar]
- Bayley P., Martin S., Jones G. The conformation of calmodulin: a substantial environmentally sensitive helical transition in Ca4-calmodulin with potential mechanistic function. FEBS Lett. 1988 Sep 26;238(1):61–66. doi: 10.1016/0014-5793(88)80225-4. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Burz D. S., Beckett D., Benson N., Ackers G. K. Self-assembly of bacteriophage lambda cI repressor: effects of single-site mutations on the monomer-dimer equilibrium. Biochemistry. 1994 Jul 19;33(28):8399–8405. doi: 10.1021/bi00194a003. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya R., Meador W. E., Means A. R., Quiocho F. A. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992 Dec 20;228(4):1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M. Young Investigator Award Lecture. Structures of larger proteins, protein-ligand and protein-DNA complexes by multidimensional heteronuclear NMR. Protein Sci. 1994 Mar;3(3):372–390. doi: 10.1002/pro.5560030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook W. J., Sack J. S. Preparation of calmodulin crystals. Methods Enzymol. 1983;102:143–147. doi: 10.1016/s0076-6879(83)02015-7. [DOI] [PubMed] [Google Scholar]
- Cook W. J., Walter L. J., Walter M. R. Drug binding by calmodulin: crystal structure of a calmodulin-trifluoperazine complex. Biochemistry. 1994 Dec 27;33(51):15259–15265. doi: 10.1021/bi00255a006. [DOI] [PubMed] [Google Scholar]
- Cox J. A. Interactive properties of calmodulin. Biochem J. 1988 Feb 1;249(3):621–629. doi: 10.1042/bj2490621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivici A., Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- Crouch T. H., Klee C. B. Positive cooperative binding of calcium to bovine brain calmodulin. Biochemistry. 1980 Aug 5;19(16):3692–3698. doi: 10.1021/bi00557a009. [DOI] [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Dedman J. R., Potter J. D., Jackson R. L., Johnson J. D., Means A. R. Physicochemical properties of rat testis Ca2+-dependent regulator protein of cyclic nucleotide phosphodiesterase. Relationship of Ca2+-binding, conformational changes, and phosphodiesterase activity. J Biol Chem. 1977 Dec 10;252(23):8415–8422. [PubMed] [Google Scholar]
- Finn B. E., Evenäs J., Drakenberg T., Waltho J. P., Thulin E., Forsén S. Calcium-induced structural changes and domain autonomy in calmodulin. Nat Struct Biol. 1995 Sep;2(9):777–783. doi: 10.1038/nsb0995-777. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J. G., Bloomfield V. A. Hydrodynamic properties of complex, rigid, biological macromolecules: theory and applications. Q Rev Biophys. 1981 Feb;14(1):81–139. doi: 10.1017/s0033583500002080. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J., Navarro S., Lopez Martinez M. C., Diaz F. G., Lopez Cascales J. J. HYDRO: a computer program for the prediction of hydrodynamic properties of macromolecules. Biophys J. 1994 Aug;67(2):530–531. doi: 10.1016/S0006-3495(94)80512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S. E., Su Z., Fan D., Means A. R. Calmodulin-cardiac troponin C chimeras. Effects of domain exchange on calcium binding and enzyme activation. J Biol Chem. 1993 Nov 25;268(33):25213–25220. [PubMed] [Google Scholar]
- Heidorn D. B., Trewhella J. Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry. 1988 Feb 9;27(3):909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- Henn S. W., Ackers G. K. Molecular sieve studies of interacting protein systems. V. Association of subunits of D-amino acid oxidase apoenzyme. Biochemistry. 1969 Sep;8(9):3829–3838. doi: 10.1021/bi00837a049. [DOI] [PubMed] [Google Scholar]
- Horiike K., Tojo H., Yamano T., Nozaki M. Interpretation of the stokes radius of macromolecules determined by gel filtration chromatography. J Biochem. 1983 Jan;93(1):99–106. doi: 10.1093/oxfordjournals.jbchem.a134183. [DOI] [PubMed] [Google Scholar]
- Ikura M., Hiraoki T., Hikichi K., Mikuni T., Yazawa M., Yagi K. Nuclear magnetic resonance studies on calmodulin: calcium-induced conformational change. Biochemistry. 1983 May 10;22(10):2573–2579. doi: 10.1021/bi00279a039. [DOI] [PubMed] [Google Scholar]
- Ikura M., Minowa O., Hikichi K. Hydrogen bonding in the carboxyl-terminal half-fragment 78-148 of calmodulin as studied by two-dimensional nuclear magnetic resonance. Biochemistry. 1985 Jul 30;24(16):4264–4269. doi: 10.1021/bi00337a002. [DOI] [PubMed] [Google Scholar]
- Ikura M., Spera S., Barbato G., Kay L. E., Krinks M., Bax A. Secondary structure and side-chain 1H and 13C resonance assignments of calmodulin in solution by heteronuclear multidimensional NMR spectroscopy. Biochemistry. 1991 Sep 24;30(38):9216–9228. doi: 10.1021/bi00102a013. [DOI] [PubMed] [Google Scholar]
- Kataoka M., Head J. F., Persechini A., Kretsinger R. H., Engelman D. M. Small-angle X-ray scattering studies of calmodulin mutants with deletions in the linker region of the central helix indicate that the linker region retains a predominantly alpha-helical conformation. Biochemistry. 1991 Feb 5;30(5):1188–1192. doi: 10.1021/bi00219a004. [DOI] [PubMed] [Google Scholar]
- Kataoka M., Head J. F., Seaton B. A., Engelman D. M. Melittin binding causes a large calcium-dependent conformational change in calmodulin. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6944–6948. doi: 10.1073/pnas.86.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka M., Head J. F., Vorherr T., Krebs J., Carafoli E. Small-angle X-ray scattering study of calmodulin bound to two peptides corresponding to parts of the calmodulin-binding domain of the plasma membrane Ca2+ pump. Biochemistry. 1991 Jun 25;30(25):6247–6251. doi: 10.1021/bi00239a024. [DOI] [PubMed] [Google Scholar]
- Kita Y., Arakawa T., Lin T. Y., Timasheff S. N. Contribution of the surface free energy perturbation to protein-solvent interactions. Biochemistry. 1994 Dec 20;33(50):15178–15189. doi: 10.1021/bi00254a029. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Vanaman T. C. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Dalgarno D. C., Levine B. A., Williams R. J. 1H-NMR studies of calmodulin. The nature of the Ca2+-dependent conformational change. Eur J Biochem. 1984 Feb 15;139(1):109–114. doi: 10.1111/j.1432-1033.1984.tb07983.x. [DOI] [PubMed] [Google Scholar]
- Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973 May 10;248(9):3313–3326. [PubMed] [Google Scholar]
- Kretsinger R. H. The linker of calmodulin--to helix or not to helix. Cell Calcium. 1992 Jun-Jul;13(6-7):363–376. doi: 10.1016/0143-4160(92)90050-3. [DOI] [PubMed] [Google Scholar]
- Kuboniwa H., Tjandra N., Grzesiek S., Ren H., Klee C. B., Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995 Sep;2(9):768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- Kung C., Preston R. R., Maley M. E., Ling K. Y., Kanabrocki J. A., Seavey B. R., Saimi Y. In vivo Paramecium mutants show that calmodulin orchestrates membrane responses to stimuli. Cell Calcium. 1992 Jun-Jul;13(6-7):413–425. doi: 10.1016/0143-4160(92)90054-v. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D., Jr, Kauzmann W. Hydration of proteins and polypeptides. Adv Protein Chem. 1974;28:239–345. doi: 10.1016/s0065-3233(08)60232-6. [DOI] [PubMed] [Google Scholar]
- Kupke D. W. Sequential volume changes on a small sample by density. Anal Biochem. 1986 Nov 1;158(2):463–468. doi: 10.1016/0003-2697(86)90576-2. [DOI] [PubMed] [Google Scholar]
- LaPorte D. C., Wierman B. M., Storm D. R. Calcium-induced exposure of a hydrophobic surface on calmodulin. Biochemistry. 1980 Aug 5;19(16):3814–3819. doi: 10.1021/bi00557a025. [DOI] [PubMed] [Google Scholar]
- Lu K. P., Means A. R. Regulation of the cell cycle by calcium and calmodulin. Endocr Rev. 1993 Feb;14(1):40–58. doi: 10.1210/edrv-14-1-40. [DOI] [PubMed] [Google Scholar]
- Matsushima N., Izumi Y., Matsuo T., Yoshino H., Ueki T., Miyake Y. Binding of both Ca2+ and mastoparan to calmodulin induces a large change in the tertiary structure. J Biochem. 1989 Jun;105(6):883–887. doi: 10.1093/oxfordjournals.jbchem.a122773. [DOI] [PubMed] [Google Scholar]
- Maune J. F., Klee C. B., Beckingham K. Ca2+ binding and conformational change in two series of point mutations to the individual Ca(2+)-binding sites of calmodulin. J Biol Chem. 1992 Mar 15;267(8):5286–5295. [PubMed] [Google Scholar]
- Mehler E. L., Pascual-Ahuir J. L., Weinstein H. Structural dynamics of calmodulin and troponin C. Protein Eng. 1991 Aug;4(6):625–637. doi: 10.1093/protein/4.6.625. [DOI] [PubMed] [Google Scholar]
- Pedigo S., Shea M. A. Quantitative endoproteinase GluC footprinting of cooperative Ca2+ binding to calmodulin: proteolytic susceptibility of E31 and E87 indicates interdomain interactions. Biochemistry. 1995 Jan 31;34(4):1179–1196. doi: 10.1021/bi00004a011. [DOI] [PubMed] [Google Scholar]
- Persechini A., Kretsinger R. H. The central helix of calmodulin functions as a flexible tether. J Biol Chem. 1988 Sep 5;263(25):12175–12178. [PubMed] [Google Scholar]
- Potschka M. Universal calibration of gel permeation chromatography and determination of molecular shape in solution. Anal Biochem. 1987 Apr;162(1):47–64. doi: 10.1016/0003-2697(87)90009-1. [DOI] [PubMed] [Google Scholar]
- Putkey J. A., Slaughter G. R., Means A. R. Bacterial expression and characterization of proteins derived from the chicken calmodulin cDNA and a calmodulin processed gene. J Biol Chem. 1985 Apr 25;260(8):4704–4712. [PubMed] [Google Scholar]
- Rasmussen C. D., Means A. R. Calmodulin is required for cell-cycle progression during G1 and mitosis. EMBO J. 1989 Jan;8(1):73–82. doi: 10.1002/j.1460-2075.1989.tb03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C. D., Means R. L., Lu K. P., May G. S., Means A. R. Characterization and expression of the unique calmodulin gene of Aspergillus nidulans. J Biol Chem. 1990 Aug 15;265(23):13767–13775. [PubMed] [Google Scholar]
- SQUIRE P. G. A RELATIONSHIP BETWEEN THE MOLECULAR WEIGHTS OF MACROMOLECULES AND THEIR ELUTION VOLUMES BASED ON A MODEL FOR SEPHADEX GEL FILTRATION. Arch Biochem Biophys. 1964 Sep;107:471–478. doi: 10.1016/0003-9861(64)90303-0. [DOI] [PubMed] [Google Scholar]
- Seamon K. B. Calcium- and magnesium-dependent conformational states of calmodulin as determined by nuclear magnetic resonance. Biochemistry. 1980 Jan 8;19(1):207–215. doi: 10.1021/bi00542a031. [DOI] [PubMed] [Google Scholar]
- Seaton B. A., Head J. F., Engelman D. M., Richards F. M. Calcium-induced increase in the radius of gyration and maximum dimension of calmodulin measured by small-angle X-ray scattering. Biochemistry. 1985 Nov 19;24(24):6740–6743. doi: 10.1021/bi00345a002. [DOI] [PubMed] [Google Scholar]
- Shea M. A., Verhoeven A. S., Pedigo S. Calcium-induced interactions of calmodulin domains revealed by quantitative thrombin footprinting of Arg37 and Arg106. Biochemistry. 1996 Mar 5;35(9):2943–2957. doi: 10.1021/bi951934g. [DOI] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966 Feb 7;112(2):346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Small E. W., Anderson S. R. Fluorescence anisotropy decay demonstrates calcium-dependent shape changes in photo-cross-linked calmodulin. Biochemistry. 1988 Jan 12;27(1):419–428. doi: 10.1021/bi00401a063. [DOI] [PubMed] [Google Scholar]
- Squire P. G., Himmel M. E. Hydrodynamics and protein hydration. Arch Biochem Biophys. 1979 Aug;196(1):165–177. doi: 10.1016/0003-9861(79)90563-0. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., James M. N. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C., Nozaki Y., Reynolds J. A., Makino S. Molecular characterization of proteins in detergent solutions. Biochemistry. 1974 May 21;13(11):2369–2376. doi: 10.1021/bi00708a021. [DOI] [PubMed] [Google Scholar]
- Taylor D. A., Sack J. S., Maune J. F., Beckingham K., Quiocho F. A. Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-A resolution. J Biol Chem. 1991 Nov 15;266(32):21375–21380. doi: 10.2210/pdb4cln/pdb. [DOI] [PubMed] [Google Scholar]
- Tjandra N., Kuboniwa H., Ren H., Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur J Biochem. 1995 Jun 15;230(3):1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- Török K., Lane A. N., Martin S. R., Janot J. M., Bayley P. M. Effects of calcium binding on the internal dynamic properties of bovine brain calmodulin, studied by NMR and optical spectroscopy. Biochemistry. 1992 Apr 7;31(13):3452–3462. doi: 10.1021/bi00128a020. [DOI] [PubMed] [Google Scholar]
- Urbauer J. L., Short J. H., Dow L. K., Wand A. J. Structural analysis of a novel interaction by calmodulin: high-affinity binding of a peptide in the absence of calcium. Biochemistry. 1995 Jun 27;34(25):8099–8109. doi: 10.1021/bi00025a016. [DOI] [PubMed] [Google Scholar]
- Wang C. L. A note on Ca2+ binding to calmodulin. Biochem Biophys Res Commun. 1985 Jul 16;130(1):426–430. doi: 10.1016/0006-291x(85)90434-6. [DOI] [PubMed] [Google Scholar]
- Warshaw H. S., Ackers G. K. Molecular sieve studies of interacting protein systems. 8. Critical evaluation of the equilibrium saturation technique using stacked gel columns. Anal Biochem. 1971 Aug;42(2):405–421. doi: 10.1016/0003-2697(71)90055-8. [DOI] [PubMed] [Google Scholar]
- Weinstein H., Mehler E. L. Ca(2+)-binding and structural dynamics in the functions of calmodulin. Annu Rev Physiol. 1994;56:213–236. doi: 10.1146/annurev.ph.56.030194.001241. [DOI] [PubMed] [Google Scholar]
- Yao Y., Schöneich C., Squier T. C. Resolution of structural changes associated with calcium activation of calmodulin using frequency domain fluorescence spectroscopy. Biochemistry. 1994 Jun 28;33(25):7797–7810. doi: 10.1021/bi00191a007. [DOI] [PubMed] [Google Scholar]
- Yazawa M., Matsuzawa F., Yagi K. Inter-domain interaction and the structural flexibility of calmodulin in the connecting region of the terminal two domains. J Biochem. 1990 Feb;107(2):287–291. doi: 10.1093/oxfordjournals.jbchem.a123040. [DOI] [PubMed] [Google Scholar]
- Yoshino H., Minari O., Matsushima N., Ueki T., Miyake Y., Matsuo T., Izumi Y. Calcium-induced shape change of calmodulin with mastoparan studied by solution X-ray scattering. J Biol Chem. 1989 Nov 25;264(33):19706–19709. [PubMed] [Google Scholar]
- Yoshino H., Wakita M., Izumi Y. Calcium-dependent changes in structure of calmodulin with substance P. J Biol Chem. 1993 Jun 5;268(16):12123–12128. [PubMed] [Google Scholar]
- Zhang M., Tanaka T., Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol. 1995 Sep;2(9):758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- le Maire M., Viel A., Møller J. V. Size exclusion chromatography and universal calibration of gel columns. Anal Biochem. 1989 Feb 15;177(1):50–56. doi: 10.1016/0003-2697(89)90012-2. [DOI] [PubMed] [Google Scholar]