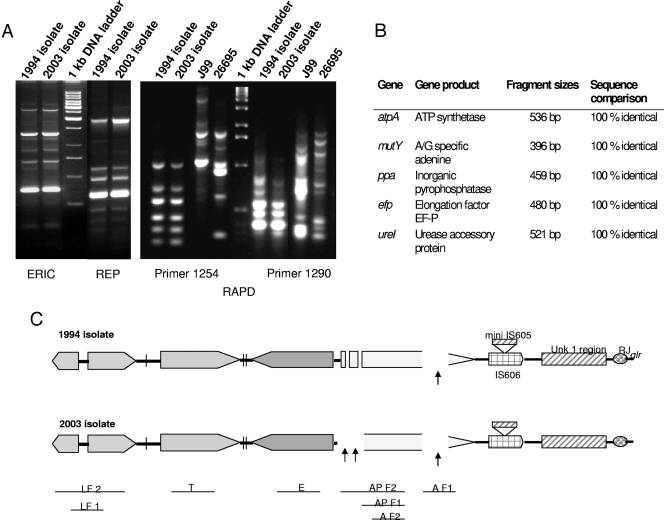
FIG. 1.
(A) ERIC, REP, and RAPD analyses of the two isolates; (B) multilocus sequence typing analysis revealing 100% identical sequences of the 5 housekeeping genes, indicating the undisputed descent of the original strain; (C) PCR-based analysis of insertion, deletion, and substitution polymorphisms within and at the extreme right junction of the cag PAI. Precise locations of rearrangements have been marked with arrows. LF 1, left end of the cag PCR product 1 (384 bp); LF 2, left end of the cag PCR product 2 (877 bp); T, cagT gene (301 bp); E, cagE gene (329 bp); AP F1, cagA promoter PCR product 1 (730 bp); AP F2, cagA promoter PCR product 2 (1,181 bp); A F1, cagA gene PCR product 1 (349 bp); A F2, cagA gene PCR product 2 (701 bp); Unk 1, unknown region 1 as described by Kersulyte et al. (17); RJ, right junction of cagA; glr, glutamate racemase.