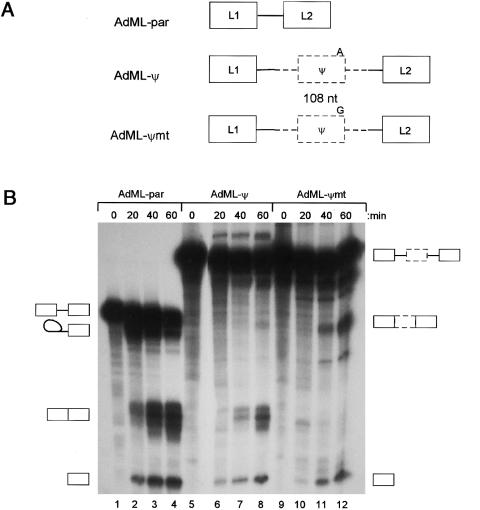
Figure 3.
Pseudoexon inclusion caused by the A→G mutation. A, Structure of the three pre-mRNA substrates used for in vitro splicing experiments. Solid boxes and lines indicate AdML sequences, and dashed boxes and lines are pseudoexon fragments and flanking intron. B, In vitro splicing. PCR fragments containing wild-type and mutant pseudoexons and surrounding introns were cloned into pCR Blunt (Invitrogen), sequenced, and used as further templates for PCR. AdML-ψ and AdML-ψmt were made by overlap-extension proof-reading PCR, using primers GHR-pe-AdMLpar2-S (5′-ccctcactaaagCAGGAGTATCATGCTGCT) and GHR-pe-AdMLpar2-A (5′-cttgactactgcTAATGACAAAATTGGCATCT), and primers AdMLpar-int51-S (5′-GCAGTAGTCAA) and AdMLpar-L2-A (5′-ATCCAAGAGTACTGGAA), and a second-round PCR with GHR-pe-AdMLpar2-S and AdMLpar-L2-A. The second-round PCR fragment was used as a megaprimer in a third-round PCR with a T7 primer, using AdMLpar as a template. Assembled AdML-ψ and AdML-ψmt PCR products were gel purified, cloned into pCR-blunt, and sequenced. Transcription templates were generated by a proof-reading PCR, with a T7 primer and AdMLpar-L2-A, and capped pre-mRNA transcribed with T7 RNA polymerase in the presence of [32P]-labeled GTP. Splicing reactions used 20 fmol of RNA, 32% Hela nuclear extract, 0.5 mM ATP, 3.2 mM MgCl2, 2.6% polyvinyl alcohol, and 60 mM KCl at 30°C (Chew et al. 2000). Reactions were deproteinized with phenol, precipitated with ethanol, and run on polyacrylamide gels ranging from 5.5% to 8%, before autoradiography. A representative in vitro splicing experiment is shown. Pre-mRNA substrates are indicated above the lanes, together with the time of the splicing reactions. The identity of the bands is indicated by the symbols to the sides of the gel and was confirmed by comparison with AdMLpar, by electrophoresis on higher-percentage gels to shift the lariat-intermediate, and by isolation of the mRNA from the gel, RT-PCR, and sequencing.