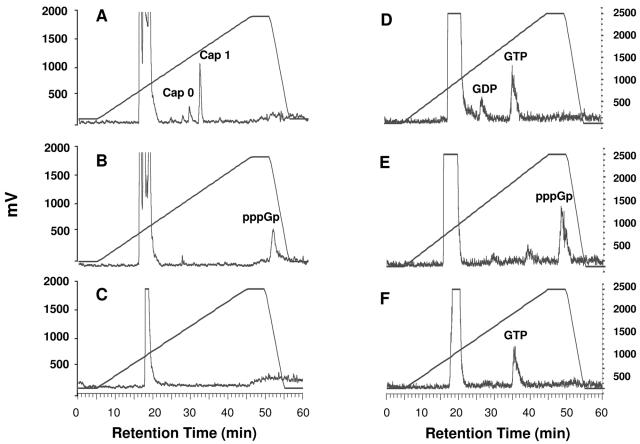
FIG. 5.
RSV polymerase inhibitors prevent RSV mRNA guanylylation. RSV transcripts were labeled with [α-32P]GTP in the presence or absence of inhibitor (compound E), digested with nuclease, and analyzed by HPLC as described in the legend of Fig. 4, except that the potassium phosphate gradient used in these experiments (and in those shown in Fig. 6) was 4 to 1,000 mM, instead of 4 to 800 mM. (A) RNase T2 digestion of RSV mRNA in absence of inhibitor. (B) RNase T2 digestion of RSV mRNA in presence of inhibitor. (C) RNase T2 and CIP treatment of RSV mRNA in presence of inhibitor. (D) NP1 digestion of RSV mRNA in presence of inhibitor. (E) T7 polymerase synthesized-NS2 RNA after RNase T2 digestion. (F) T7 polymerase-synthesized NS2 RNA after NP1 digestion. Cap 0 represents 7mGpppGp and cap 1 represents 7mGpppGmpGp. Note that although other inhibitors were more potent, compound E was used in this experiment due to better compound solubility. The y axis, in units of mV, is a measure of radioactivity by the Berthold monitor, which converts radioisotope decay to pulses of electricity (1,000 mV is approximately 8,500 cpm). The retention time of the peak in panel B varied from 48.3 to 50.9 min over seven experiments, whereas the retention time of the peak in panel E varied from 48 to 49 min.