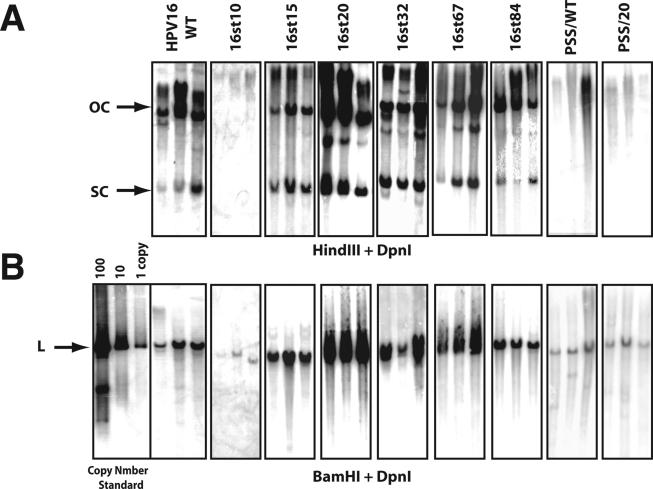
FIG. 2.
Southern analysis of total genomic DNA isolated from NIKS cell populations transfected with wild-type or E4 mutant HPV16 genomes. Shown are Storm phosphorimages of hybridization patterns from Southern analyses carried out to assess the presence of extrachromosomal HPV16 genomes (A) and the copy number of the HPV16 genomes (B) present in NIKS populations transfected with the wild-type or E4 mutant HPV16 genomes. Total genomic DNAs were extracted from NIKS cell populations at passage 1, following G418 selection. Bacterially synthesized DNA was added in each extraction as a spike DNA to ensure complete digestion of DpnI (see Materials and Methods); Southern blots for DpnI-undigested DNA are not shown. Equal amounts of total genomic DNA from each population were digested with DpnI and HindIII (no site in HPV16 genome) (A) or BamHI (one site in HPV16 genome) (B) and subjected to Southern blot analysis using a radiolabeled DNA probe for HPV16 DNA. Arrows indicate OC, SC, and linear (L) HPV16 genomes. BamHI-digested parental pEFHPV16-W12E was used as a copy number standard.