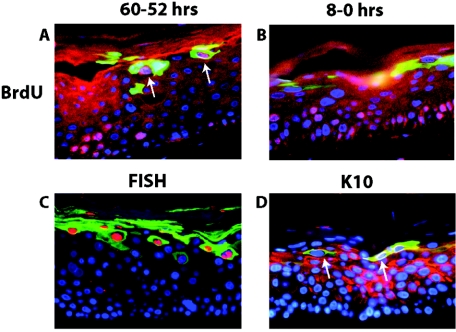
FIG. 8.
Colocalization analysis of E1∧E4 with hallmarks of the productive stage of the viral life cycle. Shown are representative cross sections of raft cultures harboring the wild-type HPV16 genome that have been subjected to double immunofluorescence to detect the relative position of E1∧E4-positive cells in relation to cells supporting DNA synthesis (A and B), viral DNA amplification (C), and aberrant differentiation (D). (A and B) Raft cultures harboring wild-type HPV16 genomes were incubated in medium containing BrdU from 60 h to 52 h prior to harvest and then cultured in medium without BrdU until harvesting (A) or from 8 h to 0 h prior to harvest (B) (see Materials and Methods). Histological cross sections of rafts were subjected to double immunofluorescence using antibodies to BrdU (red) and E1∧E4 (green) and counterstained with DAPI (blue). (A) Arrows indicate BrdU-E1∧E4 double-positive cells. (C) Histological cross sections of rafts harboring the wild-type HPV16 genome were subjected to HPV16-specific FISH. Following hybridization to the probe, sections were incubated with FITC-conjugated anti-E1∧E4 antibody (green), and the amplified HPV16 genomes were detected by rhodamine-conjugated anti-DIG antibody (red). (D) Histological cross sections of rafts harboring wild-type HPV16 were subjected to double immunofluorescence using antibodies to K10 (red) and E1∧E4 (green). K10-negative cells in the superficial spinous and granular layers are indicated by arrows.