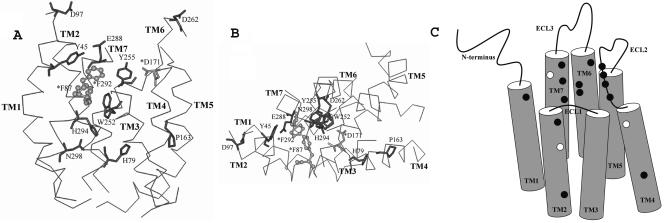
FIG. 5.
Distinct functional sites for HIV-1 gp120 and SDF-1α on a hypothetical structural model of CXCR4. The TM residues Phe87 and Phe292, required for SDF-1α binding only, are highlighted in a lighter color and represented as ball-and-stick figures with asterisks. Asp171, the only residue found to be involved in both HIV-1 gp120 and SDF-1α interactions, is highlighted in a lighter color with an asterisk. The TM residues that are involved in HIV-1 coreceptor activity only are highlighted in the darker color. Only the TM domains of CXCR4, with side (A) and top (B) views, are shown for simplicity. (C) Schematic illustration of the locations of residues found to be important for HIV-1 or SDF-1α on CXCR4 TM and ECL2 domains. The residues required for SDF-1α binding are shown as white spots, while those that are involved in HIV-1 coreceptor activity are shown as black spots. Asp171 is highlighted as a white spot.