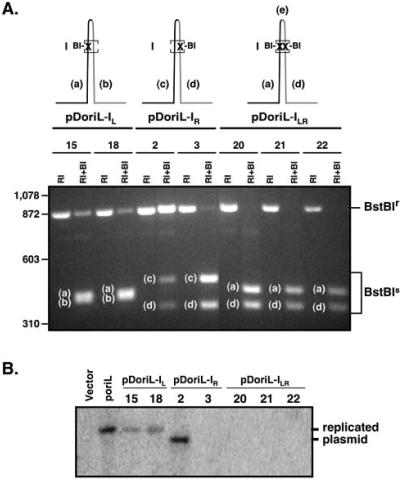
FIG. 3.
Restriction enzyme analysis and in vitro DNA replication assays with plasmids containing wild-type and mutant forms of oriL. (A) Restriction enzyme analysis of clonal isolates of oriL mutant plasmid DNA. The three mutant forms of oriL generated using the strategy outlined in Fig. 2 are diagrammed at the top of panel A. Each line represents a dsDNA oriL palindrome. “X” indicates the locations of point mutations in site I that introduce unique BstBI restriction sites. Lowercase letters in the gel photo indicate BstBI restriction fragments whose map locations are shown in the upper diagrams. Note that the small BstBI “e” fragment between the two BstBI sites in pDoriL-ILR is not detectable in the agarose gel photograph. The numbers of individual clonal isolates of each oriL type are shown under the diagrams. Plasmid DNA was cleaved with EcoRI, which liberates oriL-containing sequences from the vector (designated BstBIr), or with EcoRI and BstBI, which discriminate intact oriL sequences from mutant oriL fragments (designated BstBIs). BI, BstBI; RI, EcoRI. (B) In vitro DNA replication assay with the empty vector, vectors containing wild-type oriL, and clonal isolates of mutated forms of oriL. Vero cell monolayers were transfected with 2 μg each of the indicated plasmids. Twenty-four hours later, cells were infected at a multiplicity of 10 PFU/cell with wild-type HSV-1 strain KOS. Eighteen hours later, cells were harvested and total cellular DNA was isolated. The DNAs were digested with HindIII to linearize the plasmid and with DpnI to discriminate newly replicated plasmid DNA from input plasmid DNA. The digested DNAs were analyzed by Southern blotting using a 32P-labeled probe specific for vector sequences.