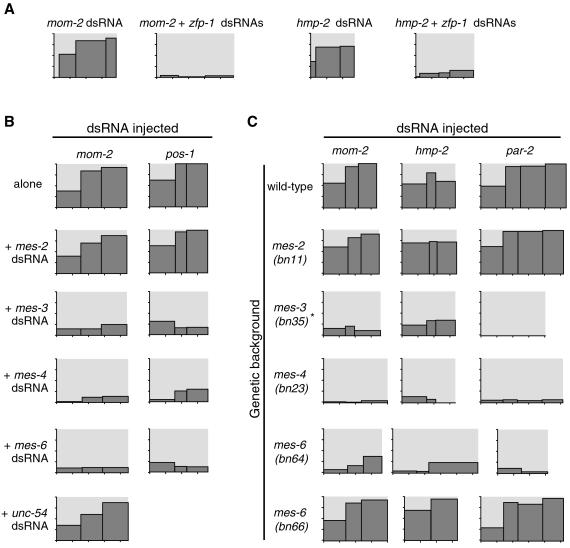
Figure 3.
Testing candidate genes for roles in RNAi. All axes are as in Fig. 1B; vertical ticks mark lethality in 25% intervals and horizontal ticks mark time at 12 h intervals. (A) Lethality produced by mom-2 dsRNA or hmp-2 dsRNA injected with or without zfp-1 dsRNA. (B) Lethality produced by mom-2 alone (top row) or coinjected with dsRNAs corresponding to mes-2, -3, -4, and -6, or a control (unc-54) dsRNA. (C) Lethality produced by injecting dsRNAs (mom-2, hmp-2, or par-2) in wild-type N2 worms or mes mutants (labeled along left side), Asterisk indicates that for par-2, bn21ts was tested at 25°C in place of bn35. ND, not determined. mes-2(bn11), mes-3(bn35), mes-4(bn23), and mes-6(bn64) (an allele with an early stop codon) each appear to be null alleles by immunostaining (refs. 36, 37, 52; Y. Fong and S. Strome, personal communication), and mes-6(bn66) (a missense allele) was found to produce small amounts of MES-6 protein by immunostaining (37). RNAi-deficient phenotypes in mes-3, -4, and -6 mutants are unlikely to be caused by genetic background, because these strains were outcrossed multiple times during construction (39), and because our results with mutants are consistent with our RNAi-to-RNAi results above. (See Fig. 5, which is published as supporting information, for complete results with additional replicates of these experiments.)