Abstract
An Escherichia coli K-12 strain was constructed with a chromosomal deletion (mutSΔ800) in the mutS gene that produced the removal of the C-terminal 53 amino acids which are not present in the MutS crystal structure. This strain has a MutS null phenotype for mutation avoidance, antirecombination, and sensitivity to cytotoxic agents in a dam mutant background.
DNA mismatch repair (MMR) plays an important role in two distinct processes, mutation avoidance and antirecombination (11, 12, 16). Mutation avoidance corrects mismatches in hemimethylated DNA behind the replication fork, and the MMR proteins MutS and MutL prevent recombination between similar but not identical (homeologous) sequences.
The crystal structure of Escherichia coli MutS bound to an oligonucleotide with a G-T mismatch has been determined using a derivative of the MutS protein, MutSΔ800, which lacks the C-terminal 53 amino acids (10). The MutSΔ800 mutant crystallizes as a dimer and retains the ability to bind DNA and ATP, just as full-length MutS (853 amino acids) does. The atomic structure of a truncated MutS from Thermus aquaticus has also been determined and is very similar to that of E. coli MutS (14). The physiological effects of the mutSΔ800 mutation have so far been studied only in multicopy (2, 4, 10), and we show below that, in single copy, it imparts a MutS null phenotype.
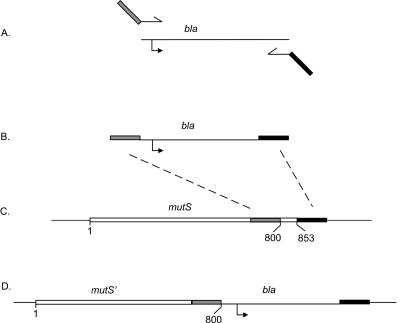
The procedure used to construct the mutSΔ800 chromosomal mutation is outlined in Fig. 1. The sequence around and including the bla gene (ampicillin resistance) and its promoter was amplified by PCR (Fig. 1A) from strain TP879 to produce a product (Fig. 1B) bearing the bla region flanked by 50-bp regions. The 5′ flanking region has the DNA sequence immediately upstream of residue 800 of mutS plus a termination codon (Fig. 1B), and the 3′ flanking region has the downstream sequence immediately following the termination codon of mutS. The PCR product was electroporated (13) into strain TP798, which constitutively expresses the products of the exo (exonuclease) and bet (beta protein) recombination genes of bacteriophage lambda (15). Recombination between the homologous regions of the PCR product and the mutS gene and its flanking sequence (Fig. 1C) produces a recombinant sequence in which the mutS gene is truncated at residue 800 and has an adjacent bla gene (Fig. 1D). By changing the upstream PCR primer sequence, we also constructed mutSΔ2, in which all but the first two and last codons of mutS were deleted, and the control mutS+ construct with the flanking bla gene.
FIG. 1.
Construction of chromosomal mutS alleles. The bla region was amplified by PCR (A) to yield a product with flanking sequences homologous to the distal end of codon 853 of the mutS gene (B). Recombination along the dotted lines between the PCR fragment and the homologous regions in the chromosome (C) yields an ampicillin-resistant truncated mutS gene at codon 800. The gray rectangle indicates 50 bp of sequence immediately upstream of codon 800, and the black rectangle indicates 50 bp of sequence immediately downstream of the mutS termination codon. The bent arrow denotes the promoter region for the bla gene.
We measured the levels of native and mutant MutS, by Western blotting (5, 7), in strains with the chromosomal constructs as well as multicopy plasmids which were in a mutS null host (Fig. 2 and Table 1). The levels in GM8311 (mutS+) were the same as those in AB1157 (mutS+) and were increased fourfold in GM7451, which harbors pMQ372 (mutS+), but no MutS was detected in GM8313 (mutSΔ2). Strain GM8315 (mutSΔ800) contained 2.5-fold less MutS than that contained by GM8311 (mutS+), but in multicopy (GM7453), the level was the same as that for the wild-type strain.
FIG. 2.
Cellular levels of MutS. The amount of MutS, as determined by Western blotting, is shown for (lane a) AB1157, wild-type control; (lane b) GM4799 (mutS458)/pMQ372 (mutS+); (lane c) GM8311 (mutS+-bla); (lane d) GM8313 (mutSΔ2-bla); (lane e) GM8315 (mutSΔ800-bla); and (lane f) GM4799 (mutS458)/pmutSΔ800. Equal amounts of cell extract were loaded in each lane. There was no detectable MutS in extracts of strain GM4799 (data not shown).
TABLE 1.
E. coli and Salmonella strains used in the study
| Strain | Description | Reference or source |
|---|---|---|
| AB259 | relA1 spoT1 thi-1 supQ80 | E. A. Adelberg |
| AB1157 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4-rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 | E. A. Adelberg |
| GM3819 | AB1157 but Δdam-16::Kan | Lab stock |
| GM4799 | AB1157 but mutS458::mTn10Kan | Lab stock |
| GM7451 | GM4799/pMQ372(mutS+) | Lab stock |
| GM7453 | GM4799/pmutSΔ800 | Lab stock |
| GM8311 | AB1157 but mutS+-blaa | Lab stock |
| GM8313 | AB1157 but mutSΔ2-blaa | Lab stock |
| GM8315 | AB1157 but mutSΔ800-blaa | Lab stock |
| GM8317 | GM3819 but mutS+-bla | Lab stock |
| GM8319 | GM3819 but mutSΔ2-bla | Lab stock |
| GM8321 | GM3819 but mutSΔ800-bla | Lab stock |
| SA534b | serA13 rfa-3058 | K. E. Sanderson |
| TP798 | MG1655 Δ(recC-ptr-recB-recD)::Ptac- gam-bet-exo-cat | A. R. Poteete et al. (15) |
| TP879 | AB1157 but Δ(recC-ptr-recB-recD)::Ptac- gam-bet-exo-pae-recA+-bla | A. R. Poteete |
Referred to as mutS+, mutSΔ2, and mutSΔ800 in the text.
Salmonella enterica serovar Typhimurium.
The strains bearing the mutSΔ2, mutSΔ800, and wild-type alleles were tested for reversions of the argE3 marker and for mutations to rifampin resistance as described elsewhere (4). The results in Table 2 show that with the wild-type strain, a low rate of resistance or reversion was observed. However, for the strains with mutSΔ2 and mutSΔ800, the number of rifampin-resistant mutants increased about 60-fold for both, and the increases for Arg+ revertants were about 60- and 40-fold, respectively.
TABLE 2.
Spontaneous mutant frequencies
| Strain | Frequency (fold increase) of:
|
|
|---|---|---|
| Rifr | Arg+ | |
| GM8311 (wild type) | 1 | 3 |
| GM8313 (mutSΔ2) | 63 | 66 |
| GM8315 (mutSΔ800) | 55 | 38 |
Each of the chromosomal mutS mutants was used as a recipient in conjugal crosses with E. coli (homologous) or Salmonella enterica serovar Typhimurium (homeologous) donors as described previously (4). For the homologous crosses with the E. coli donor, recombinants are formed at the same frequency, indicating that there is no effect on homologous recombination stemming from the construction of the chromosomal mutS mutations (Table 3). With the Salmonella donor and a wild-type mutS recipient, no recombinants were detected. In contrast, the mutSΔ2 and mutSΔ800 recipients increased the ability to form recombinants 11,000- and 27,000-fold, respectively, with the same Salmonella donor (Table 3). These increases were abrogated in crosses with recA deletion derivatives of the mutSΔ2 and mutSΔ800 mutant recipients (data not shown), indicating that the mutS mutations reduce antirecombination function in a recA-dependent manner.
TABLE 3.
Recombination frequencies in homologous and homeologous crossesa
| Donor strain | Recipient mutant | Frequency |
|---|---|---|
| AB259 | GM8311 (mutS+) | 2.0 × 106 |
| GM8313 (mutSΔ2) | 1.8 × 106 | |
| GM1315 (mutSΔ800) | 1.9 × 106 | |
| SA534 | GM8311 (mutS+) | <1 |
| GM8313 (mutSΔ2) | 11.1 × 103 | |
| GM1315 (mutSΔ800) | 27.1 × 103 |
Thr+ Leu+ Strr Ampr recombinants were selected, and the frequency is expressed per 108 donors.
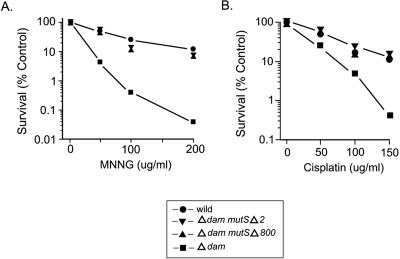
E. coli dam mutS+ mutants are more sensitive to exposure to MNNG (N-methyl-N′-nitro-N-nitrosoguanidine) (Fig. 3A) and cisplatin (Fig. 3B) than the wild type (6, 8, 9) is, but the dam mutSΔ2 deletion strain is as resistant to both treatments as the wild type is (Fig. 3), based on determinations using the protocol described previously (4). Figure 3 also shows that dam mutSΔ800 bacteria are as resistant to MNNG and cisplatin as the dam mutSΔ2 cells are.
FIG. 3.
Cell survival after exposure to (A) MNNG and (B) cisplatin. Cells in the logarithmic phase of growth were exposed to MNNG for 10 min or cisplatin for 60 min, and survival levels were determined as a function of dose. Circles, wild type (GM8311); inverted triangles, dam mutSΔ2 mutant (GM8319); triangles, dam mutSΔ800 mutant (GM8321); squares, dam mutant (GM3819).
We conclude that the mutSΔ800 mutation in a single copy on the chromosome confers a mutS null phenotype to a cell to the same degree as the mutSΔ2 deletion mutation does for mutation avoidance, antirecombination, and resistance to cytotoxic agents. On a multicopy plasmid, the mutSΔ800 mutation in a dam mutS host confers a “split” phenotype, where mutation avoidance (2, 4, 10) and MNNG sensitivity are at the wild-type levels but antirecombination and resistance to cisplatin are severely diminished (4).
The MutS null phenotype of the mutSΔ800 strain is due in part to the decreased cellular level of MutSΔ800 compared to that of MutS (Fig. 2), indicating that the C-terminal 53 amino acids impart stability to the protein. Even when corrected so that the levels of the proteins are similar, as in strains with the multicopy plasmid mutSΔ800 and single-copy mutS, there is still the deficiency of antirecombination and resistance to cisplatin (4). Furthermore, purified MutSΔ800 protein has a lower affinity than MutS does for certain oligonucleotides with base pair mismatches and, in the presence of other MMR components, reduces the efficiency of MutH-induced incision at hemimethylated GATC sequences in vitro (3). The lower cellular amount of MutSΔ800 protein, therefore, cannot be the sole explanation for the phenotypic differences between wild-type and mutSΔ800 strains.
The data presented here indicate that the C-terminal 53 amino acids are essential for MutS function in vivo. At present, the only known feature associated with this region comes from equilibrium sedimentation and gel filtration studies showing that MutS dimers can assemble into higher-order oligomeric structures, while MutSΔ800 is restricted to dimer formation only (3). A similar oligomeric composition occurs with the MutS protein from Thermus species (1). At present, the location of the tetramerization sequence is not known, but we are currently attempting to localize it.
Acknowledgments
We thank Tony Poteete for assistance in designing the chromosomal mutS mutation protocol and Jennifer Saporita and Mary Munson for technical advice regarding antibody purification and Western blots.
This work was supported by grant GM63790 from the National Institutes of Health.
REFERENCES
- 1.Biswas, I., C. Ban, K. G. Fleming, J. Qin, J. W. Lary, D. A. Yphantis, W. Yang, and P. Hsieh. 1999. Oligomerization of a MutS mismatch repair protein from Thermus aquaticus. J. Biol. Chem. 274:23673-23678. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, I., G. Obmolova, M. Takahashi, A. Herr, M. A. Newman, W. Yang, and P. Hsieh. 2001. Disruption of the helix-u-turn-helix motif of MutS protein: loss of subunit dimerization, mismatch binding and ATP hydrolysis. J. Mol. Biol. 305:805-816. [DOI] [PubMed] [Google Scholar]
- 3.Bjornson, K. P., L. J. Blackwell, H. Sage, C. Baitinger, D. Allen, and P. Modrich. 2003. Assembly and molecular activities of the MutS tetramer. J. Biol. Chem. 278:34667-34673. [DOI] [PubMed] [Google Scholar]
- 4.Calmann, M. A., A. Nowosielska, and M. G. Marinus. 2005. Separation of mutation avoidance and antirecombination functions in an Escherichia coli mutS mutant. Nucleic Acids Res. 33:1193-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng, G., H.-C. T. Tsui, and M. E. Winkler. 1996. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 178:2388-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fram, R. J., P. S. Cusick, J. M. Wilson, and M. G. Marinus. 1985. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol. Pharmacol. 28:51-55. [PubMed] [Google Scholar]
- 7.Gage, S. D., and W. R. Kobertz. 2004. KCNE3 truncation mutants reveal a bipartite modulation of KCNQ1 K+ channels. J. Gen. Physiol. 124:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, M., and R. Wagner. 1981. N-Methyl-N′-nitro-N-nitrosoguanidine sensitivity of E. coli mutants deficient in DNA methylation and mismatch repair. Mol. Gen. Genet. 184:562-563. [DOI] [PubMed] [Google Scholar]
- 9.Karran, P., and M. G. Marinus. 1982. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature 296:868-869. [DOI] [PubMed] [Google Scholar]
- 10.Lamers, M. H., A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Wind, and T. K. Sixma. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407:711-717. [DOI] [PubMed] [Google Scholar]
- 11.Marti, T. M., C. Kunz, and O. Fleck. 2002. DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 191:28-41. [DOI] [PubMed] [Google Scholar]
- 12.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 13.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obmolova, G., C. Ban, P. Hsieh, and W. Yang. 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407:703-710. [DOI] [PubMed] [Google Scholar]
- 15.Poteete, A. R., A. C. Fenton, and A. Nadkarni. 2004. Chromosomal duplications and cointegrates generated by the bacteriophage lambda Red system in Escherichia coli K-12. BMC Mol. Biol. 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schofield, M. J., and P. Hsieh. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579-608. [DOI] [PubMed] [Google Scholar]