Abstract
Mutations of genes needed for melanocyte function can result in oculocutaneous albinism. Examination of similarities in human gene expression patterns by using microarray analysis reveals that RAB38, a small GTP binding protein, demonstrates a similar expression profile to melanocytic genes. Comparative genomic analysis localizes human RAB38 to the mouse chocolate (cht) locus. A G146T mutation occurs in the conserved GTP binding domain of RAB38 in cht mice. Rab38cht/Rab38cht mice exhibit a brown coat similar in color to mice with a mutation in tyrosinase-related protein 1 (Tyrp1), a mouse model for oculocutaneous albinism. The targeting of TYRP1 protein to the melanosome is impaired in Rab38cht/Rab38cht melanocytes. These observations, and the fact that green fluorescent protein-tagged RAB38 colocalizes with end-stage melanosomes in wild-type melanocytes, suggest that RAB38 plays a role in the sorting of TYRP1. This study demonstrates the utility of expression profile analysis to identify mammalian disease genes.
Melanocytes are specialized pigment-producing cells that are responsible for coloration of skin, eyes, and hair. Coat color alterations resulting from melanocyte defects are easily identifiable in mice. These mouse mutants are proving valuable for the identification of candidate human disease genes and the elucidation of mechanisms underlying cellular function. To date, there are 99 loci in the mouse that, when mutated, affect pigmentation (Dorothy C. Bennett, personal communication). However, the underlying genetic defect has been identified for only approximately one-third of these loci, leaving over 60 yet to be characterized (Mouse Genome Informatics, http://www.informatics.jax.org/).
Disorders with reduced pigmentation can be placed into two groups according to whether they affect melanocyte differentiation or melanosome function, the pigment producing organelle in the melanocyte. Examples of the first group include Piebaldism and Waardenburg syndrome, characterized by a localized absence of melanocytes resulting in “white patch” patterns. Genes affected in these disorders, KIT, MITF, PAX3, SOX10, EDNRB, EDN3, are involved in specification, migration, and survival of the melanocyte lineage (1). Mouse models of these disorders have characteristic spotted coat patterns (1). Oculocutaneous albinism (OCA) I-IV, Chediak–Higashi syndrome (CHS), Hermansky–Pudlak syndrome (HPS) I-III, and Griscelli syndrome (GS) correspond to the second group. The molecular defects contributing to the reduced pigmentation in OCA occurs in genes (TYR, TYRP1, P, and AIM1) that affect mainly melanosome formation and the amount and type of melanin pigment formed (2, 3). Genes responsible for HPS, CHS, and GS are involved in the regulation of vesicle traffic including melanosomes within in the cell, and include HPS1, AP3, HPS3, CHS1, MYO5A, and RAB27A (1, 4).
Given the genetic heterogeneity of pigmentation disorders in both mouse and human, we sought to identify additional candidate disease genes by using cDNA microarrays (5) to examine the expression behavior of genes over a variety of neural crest-derived and other control cell lines. Clustering of the resulting expression profiles provided a powerful way to organize the common patterns found among thousands of gene expression measurements and identify genes with similar distinctive expression patterns among the experimental samples (6). Analysis of genes contained within a cluster has revealed that these genes are often functionally related within the cell (7, 8). Using this approach we identified genes clustered with known pigmentation genes, thereby categorizing RAB38 as a candidate pigmentation gene. Further analysis confirms that RAB38 is a melanosomal protein, mutated in the mouse pigmentation mutant, chocolate (cht), and important for the sorting of the melanosomal protein tyrosinase-related protein 1 (TYRP1) in melanocytes.
Materials and Methods
Cell Culture.
All cells for microarray analysis were grown to 90% confluence at 37°C, 5% CO2. Melanoma cell lines were grown as described (9). 293T, U138, and HeLa cells were grown in DMEM containing 10% FBS, 2 mM l-glutamine, and 100 units/ml each penicillin and streptomycin. Primary melanocyte cultures were grown as described (10). Melan-a cells were cultured in RPMI 1640 media containing 10% FBS, 2 mM l-glutamine, 10 mM sodium pyruvate, 100 units/ml each penicillin and streptomycin, 200 nm phorbol 12-myristate 13-acetate, 0.01 mM sodium bicarbonate, and 0.1 mM 2-mercaptoethanol at 5% CO2.
RNA Preparation.
Cells for microarray analysis were obtained in pools of four 500-cm2 dishes, harvested by scraping, washed in PBS, and pelleted. Pellets were lysed in 10 ml Trizol reagent (Invitrogen). Two milliliters of chloroform was added, and the sample was shaken and centrifuged to separate phases. The aqueous layer was removed and an equal volume of 75% ethanol was added dropwise while vortexing. Sample was applied to a RNeasy maxi column (Qiagen, Valencia, CA) and purification protocol was followed. Samples were eluted in water, precipitated with 3 M sodium acetate, and stored at −80°C. RNA pellets were resuspended in diethyl pyrocarbonate-treated water to 1 μg/μl concentration and concentrated on a Microcon 100 column to 7–10 μg/μl.
Labeling and Hybridization.
RNA was reverse-transcribed to fluorescent-labeled cDNA and cohybridized on slides in experimental/reference pairs. Expressed sequence tag clone inserts applied to slides were prepared as described (11). Reverse transcription fluorochrome-labeled cDNA was generated as described at http://www.nhgri.nih.gov/DIR/Microarray/main.html. For reactions, 60 μg of total RNA (Cy3) or 120 μg of total RNA (Cy5) was used. Hybridizations were carried out in a volume of 40 μl at 65°C in a humidified chamber for 16 h. Slides were washed at room temperature in 0.5 × SSC/0.1% SDS for 3 min, then in 0.6 × SSC for 3 min. Slides were immediately spun dry by centrifugation.
Image Acquisition and Analysis.
Fluorescence signal intensities for cy3 (532λ) and cy5 (635λ) fluorochromes were obtained with a Genepix 4000a scanner (Axon Instruments, Foster City, CA) at 10-μm resolution. A set of 88 housekeeping control genes was used for normalization of labeling efficiency (12). Expression profile analysis was performed with a clustering algorithm by using an average-linkage method and Pearson's correlation similarity measurement (http://microarray.nhgri.nih.gov/genecluster).
Organization of the Mouse RAB38 Gene.
blast was used to compare the Rab38 mRNA sequence (GenBank accession no. AY062237) against mouse genomic sequencing trace archives (www.ncbi.nlm.nih.gov/Traces/trace.cgi?). Genomic organization was confirmed by using spidey (www.ncbi.nlm.nih.gov/spidey/). Gene organization was experimentally confirmed through PCR and DNA sequencing of genomic fragments. The confirmed exon and surrounding intronic sequence have been deposited to GenBank (exon 1, accession no. AF448441; exon 2, accession no. AF448442; exon 3, accession no. AF448443).
In Situ Hybridization.
FVB/NJ mouse embryos (Jackson Laboratory) were fixed overnight in 4% paraformaldehyde in PBS. Reverse-transcribed digoxigenin-conjugated probes were made from linearized plasmids and/or PCR products with polymerase binding site linkers (all reagents from Roche Molecular Biochemicals). The following DNA sources were used for probe synthesis: tyrosinase, cDNA clone 4633402C07; Tyrp1, reverse transcription–PCR from B16F10Y total RNA (TYRP15′T3F-GCGCGAATTAACCCTCACTAAAGGGTCTGAGCACCCCTGTCTTCT TYRP15′T7RGCGCGTAATACGACTCACTATAGGGCCCAGTTGCAAAATTCCAGT); Dct, cDNA (13); AimI/Matp, RIKEN cDNA clone G370045L22; Mlsn1 reverse transcription–PCR from B16F10Y total RNA (MLSN R T7-GCGGGTAATACGACTCACTATAGGGGCCACAAACATGTCCTACTTAC; MLSN FT3GCGCGAATTAACCCTCACTAAAGGGAAGCTTCCGGACTCTCTAC); Rab38, RIKEN cDNA clone 23–10011-F14. In situ hybridizations were performed by using published protocols (14) with the following modifications. After probe hybridization, Ribonuclease A digestion was omitted, and Tris-buffered saline was used in place of PBS. BM-purple substrate (Roche, Molecular Biochemicals) was used in place of 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium.
Mutation Detection.
Mouse RAB38 primer pairs were designed to amplify the three protein-coding exons as well as a small amount of flanking intronic DNA: Rab38 Ex1F (TAGGAAGGAGGATTAAACCCG) and Rab38 Ex1R (GAACTCCTCATGGCTCACTCC), yielding a 428-bp product; Rab38 Ex2F (GGATATGAAGCTCCAGTGTAGTGTAC) and Rab38 Ex2R (CACTGGACAGAAACATTATTGTCAC), yielding a 464-bp product; and Rab38 Ex3F (AAGTTATCAGCCAGTGAGATACTGTG) and Rab38 Ex3R (CACATGTGGTATATCTATCCTGACG), yielding a 526-bp product. After separation on a 1% agarose gel, PCR products were excised and purified by using QIAquick Gel Extraction Kit (Qiagen) and directly sequenced. Mutations were confirmed by using primers designed to amplify a 213-bp fragment surrounding the G146T sequence (cht Ex1F-GGCCTCCAGGATGCAGACACC and cht Ex1R-CCAGCAATGTCCCAGAGCTGC). SexAI and BsaJI restriction enzyme digests were performed by using 10 μl of PCR product, 2.5 units enzyme (New England Biolabs) along with 1× of the supplied BSA and digest buffer. Reactions were incubated overnight at the manufacturer's suggested temperature and electrophoresed on a 2% agarose gel.
Cell Transfection.
RAB38-green fluorescent protein (GFP) constructs were generated by PCR amplifying mouse Rab38 with att site linker primers AttB1-RRab (GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGCAGACACCTCACAAG) and AttB2-RRab-STP (GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAGGATTTGGCACAGCCAGA) and Gateway cloning into pDest 53 (Invitrogen) as per the manufacturer's instructions. RAB38-GFP was transfected into melan-a cells by using Lipofectamine 2000 (Invitrogen) with a DNA/Lipofectamine 2000 ratio of 1.6 μg/4 μl in a 4-cm2 surface area as per reagent instructions. After 72 h, cells were fixed and antibody was stained. TYRP1 antibody (MEL5, Signet Laboratories, Dedham, MA) was used at a 1:200 dilution (15).
Bleeding Times.
Bleeding times were assayed in four C57BL/6J Rab38cht/Rab38cht and four C57BL/6J animals, as described (16). Assayed mice were 6–12 weeks of age.
Results
Expression Profile Analysis Identifies RAB38 as cht Candidate Gene.
To identify novel and uncharacterized genes involved in melanocyte function and disease, we generated a collection of cDNA clones to be used for expression profile and functional analyses (12). We have previously shown that cDNA clones from a human melanocyte image consortium library, 2NbHM, (http://www.ncbi.nlm.nih.gov/UniGene/lib.cgi?ORG=Hs&LID=198) are appropriate for gene expression studies aimed at understanding melanocyte development and function (5, 12). For this analysis, 4,356 nonsequence-verified cDNA clones from library 2NbHM were printed to glass slides, along with sequence-verified melanocyte control genes. Hybridization data were obtained from 17 cell lines representing neural crest- and non-neural crest-derived tissues and used for hierarchical cluster analysis. Included were 11 melanoma cell lines (9), three rhabdomyosarcoma cell lines, one glioblastoma cell line, HeLa, and 293T. Array hybridizations for each of these cell lines were performed in a pair-wise fashion, using RNA from cell line UACC903(+6) as a reference. UACC903(+6) is an amelanotic melanoma cell line rendered nontumorigenic by the introduction of a region of human chromosome 6 (17) and has been used previously for expression profile analysis of melanoma lines with a different set of cDNA clones (9).
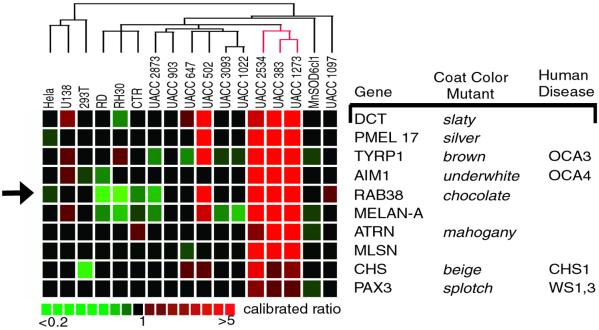
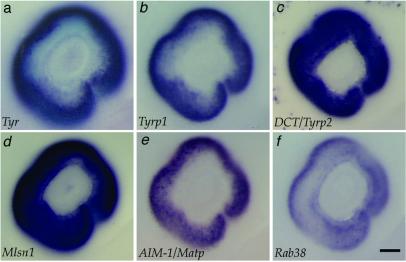
Analysis of expression profiles demonstrated that nine melanocyte control genes (DCT, TYRP1, PMEL17, AIM-1, MELAN-A/MART1, MLSN, ATRN, PAX3, and CHS1) showed similar expression variation among the samples analyzed (Fig. 1). We sought to identify candidate pigmentation genes in the 4356 cDNA set by selecting expressed sequence tag clones that exhibited similar expression variation to the melanocyte control genes. RAB38 was found to have a similar expression variation to the nine melanocyte genes (Fig. 1). As four of the nine melanocyte genes examined (TYRP1, DCT, MLSN, and AIM1) were shown to be expressed in the melanocytes of the retinal pigmented epithelium (RPE) at embryonic day 11.5 (Fig. 2) we screened candidate pigmentation genes by in situ hybridization analysis. Consistent with the placement of RAB38 within this collection of melanocyte genes, whole-mount in situ analysis demonstrated that RAB38 was also expressed in the melanocytes of the RPE at this age (Fig. 2). Previous Northern blot analyses also demonstrated restricted expression of RAB38 to melanocyte-derived cell lines (18).
Figure 1.
Microarray expression profile of melanocyte control genes and RAB38. Expression profiles of nine genes known to be involved in pigmentation are similar to the expression profile of RAB38 (arrow). Columns to the right list identified mouse and human pigmentation disorders corresponding to a given gene. Hybridized sample cell lines are listed across the top of the expression profiles. Relative gene expression is evaluated as a calibrated ratio (sample cell line/MnSOD6cl1 reference). Pseudocolor scale for ratio values is below.
Figure 2.
Rab38 expression in the RPE is similar to that of melanogenic enzymes. All panels show the eye at embryonic day 11.5. The melanogenic enzymes tyrosinase (Tyr) (a), tyrosinase-related protein 1 (Tyrp1) (b), and Dopachrome tautamerase (DCT/Tyrp2) (c) all show expression in the RPE at this developmental stage. Additional control genes from the microarray cluster data, Melastatin1 (Mlsn) (d) and AimI/Matp (e), show expression in the RPE. Rab38 is expressed in the RPE at embryonic day 11.5 (f) and at embryonic days 10.5 and 12.5 (data not shown). (Scale bar equals 100 μm.)
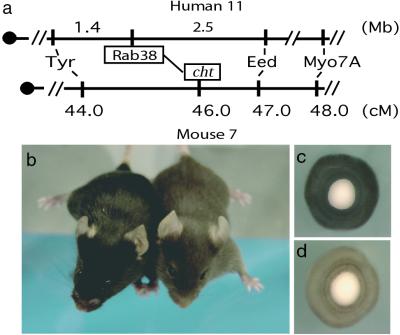
Using the recently available human genome sequence, we were able to determine that RAB38 was located on human chromosome 11, flanked proximally by TYR and distally by EED and MYO7A (Fig. 3a). A conserved linkage group on mouse chromosome 7 was identified by comparison of the human genome map with the mouse genome mapping data (http://www.informatics.jax.org/searches/homology_form.shtml). Closer analysis of loci in the mouse conserved linkage group determined that an uncloned mouse pigmentation mutant, cht, was contained within this interval (19) (Fig. 3a). The cht mutation arose as a spontaneous, isogenic mutation on an inbred C57BL/6J background and has been maintained on this background since 1984 (20). Cht/cht mice are identifiable at birth by lighter skin and eyes and at weaning by a deep-brown coat color when compared with C57BL/6J parental strain (Fig. 3 b–d). Thus, RAB38 was implicated as a candidate gene for the cht locus based on genomic map position and the expression of RAB38 in RPE and melanocytes, the cell types affected in cht/cht mice.
Figure 3.
Map location and phenotype of cht/cht mouse. (a) Human RAB38 maps 1.4 Mb distal to tyrosinase and 2.5 Mb proximal to EED genes on human chromosome 11. In the corresponding mouse chromosome 7 syntenic region, gene order is conserved. The genetic location of the cht locus maps to this same interval as RAB38. (b) Photograph of C57BL/6J+/+ (Left, black) and C57BL/6J Rab38cht/Rab38cht (Right, brown) mice. Eyes from 2-day-old mice (c) wild-type C57Bl6/J+/+ shows normal pigmentation, whereas (d) Rab38cht/Rab38cht eye shows reduced pigmentation. (Magnification: ×50.)
Mutation of RAB38 Causes Melanocyte Defects in cht/cht Mice.
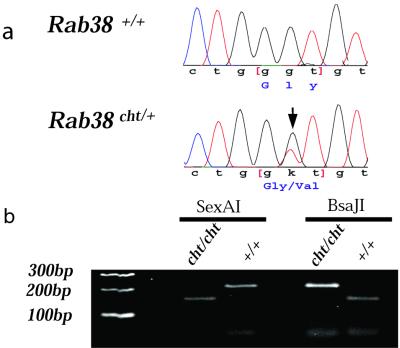
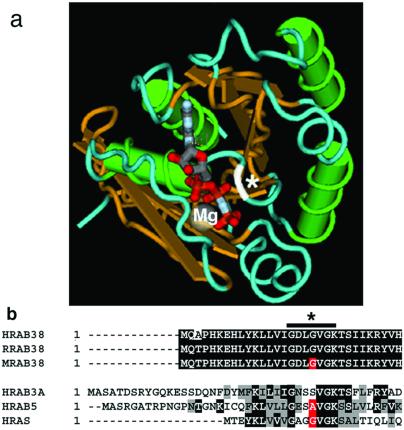
Genomic sequence flanking exon/intron boundaries for the three mouse Rab38 exons was obtained from mouse trace archive genomic sequence (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?). DNA from C57BL/6J cht/+ animals was obtained from Jackson Laboratories Mouse DNA Resource, amplified by using genomic primers, and directly sequenced. A unique G146T nucleotide mutation in exon 1 was identified in the cht allele (Fig. 4a). This sequence alteration was confirmed by restriction digest in multiple cht/cht DNA samples, as the resulting nucleotide substitution changed a BsaJI site (CCNNGG) to a SexA1 restriction site (ACCWGGT) (Fig. 4b). This sequence alteration was not detected in analysis of eight additional inbred strains (CAST/Ei, SPRET/Ei, 129/SVJ, FVB/NJ, AKR/J, A/J, DBA/1J, and BALB/cJ) (data not shown). The RAB38 protein demonstrates highly conserved amino acid identity among mammals: human/rat (96.2%), human/mouse (93.8%), rat/mouse (95.2%) (Fig. 5b). The G19V cht mutation is located within the highly conserved phosphate/Mg2+ domain and is predicted to contact GTP directly in the nucleotide binding pocket (Fig. 5a).
Figure 4.
Rab38 mutation causes the cht mouse phenotype. (a) Comparison of Rab38 sequence between wild-type C57BL/6J+/+ and C57BL/6J Rab38cht/+ DNA identified a G146T nucleotide change (arrow) for the cht allele. This nucleotide change was never seen in eight additional inbred strains analyzed (data not shown). (b) The G146T mutation creates a SexA1 restriction enzyme site in C57BL/6J Rab38cht/Rab38cht DNA and ablates a BsaJI restriction site present in wild-type Rab38 sequence. A 216-bp region surrounding the G146T nucleotide mutation was amplified from both C57BL/6J+/+ and C57BL/6J Rab38cht/Rab38cht DNA. SexA1 digests the PCR fragment of C57BL/6J Rab38cht/Rab38cht but not C57BL/6J+/+ (Left); BsaJ1 digests the PCR fragment of C57BL/6J+/+ but not C57BL/6J Rab38cht/Rab38cht (Right).
Figure 5.
RAB38 G19 is located in the GTP binding pocket. (a) The three-dimensional location of amino acid G19 of RAB38 in relation to the nucleotide binding site was determined by using the molecular modeling database (MMDB) (40) based on the crystal structure for RAB3a (MMDB 10125) (34). Overlaying RAB38 sequence with that of RAB3a sequence identified amino acid S32 of RAB3a as equivalent to G19 of RAB38. The program CN3D 3.0 was used to indicate the location of the RAB38 G19 (white), predicting interaction with the bound nucleotide. Protein structure is indicated by color: green, α helices; gold, β sheet; blue, random coils; white, site of RAB3a S32 equivalent to RAB38 G19 located at the nucleotide binding site; gray, Mg++ ion; red-gray, GppNHp nucleotide analog. (b) Alignments of highly conserved N-terminal region comparing human RAB38 (NP_071732), rat RAB38 (AAA42000), and mouse RAB38 (AK009296.1). Corresponding alignments to human RAB3a (P20336), human RAB5 (F34323) and human N-RAS (TVHURA). Sequence alignment was done by using the clustalw algorithm (41). Bars indicate highly conserved regions that occupy the nucleotide binding pocket, demonstrated in the x-ray crystal structure of RAB3a (42). Black denotes sequence identity, gray denotes sequence conservation, red denotes the conserved amino acid.
Rab38cht Results in Decreased Efficiency of Targeting Tyrp1 to End-Stage Melanosomes.
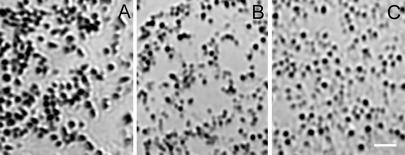
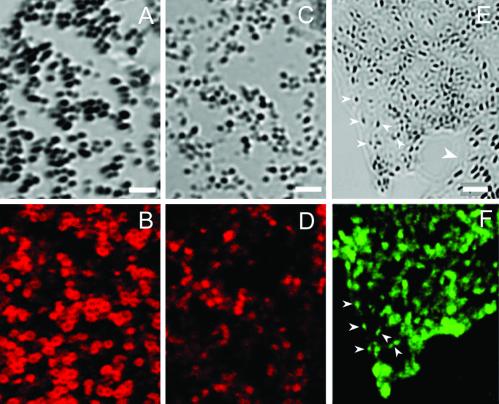
Analysis of melanocytes cultured from newborn mice revealed that C57BL/6J Rab38cht/Rab38cht melanocytes (Fig. 6B) contain small, circular melanosomes with a brown hue similar to those observed in C57BL/6J Tyrp1b/Tyrp1b melanocytes (21) (Fig. 6C), but distinct from the intensely black, oval melanosomes seen in C57BL/6J+/+ melanocyte cultures (Fig. 6A). Given that Tyrp1 mutations cause a switch from black to brown pigment in brown mice, and given that Rab GTPases play a central role in protein trafficking (22, 23), we hypothesized that the targeting of TYRP1 to the melanosome might be defective in Rab38cht/Rab38cht melanocytes. Consistent with this idea, end-stage melanosomes in the Rab38cht/Rab38cht melanocytes stain much more weakly for TYRP1 than do end-stage melanosomes in control melanocytes (Fig. 7 A–D). This observation suggests that RAB38 regulates traffic of vesicular intermediates that move TYRP1 from the trans-Golgi network to end-stage melanosomes. Consistent with this finding, GFP-tagged RAB38 colocalizes with end-stage melanosomes in wild-type cells (Fig. 7 E and F).
Figure 6.
The cht/cht melanosomes are similar in morphology to Tyrp1b melanosomes. Shown are bright-field images of melanosomes from the periphery of primary cultured melanocytes, isolated from (A) C57BL/6J+/+, (B) C57BL/6J Rab38cht/Rab38cht mice, and (C) Tyrp1b/Tyrp1b melan-b cells. Melanosomes from wild-type melanocytes are oval and darkly pigmented. In contrast, Rab38cht/Rab38cht melanosomes are smaller, more circular and less pigmented, similar to melanosomes from (C) Tyrp1b/Tyrp1b melan-b cells. (Scale bar = 2 μM.)
Figure 7.
RAB38 is a melanosomal protein needed for appropriate TYRP1 trafficking. Shown are (A and B) C57BL/6J+/+ and (C and D) C57BL/6J Rab38cht/Rab38cht primary melanocyte cultures. (A and C) Bright-field images of melanosomes in the cell periphery with matching confocal images of identical exposure demonstrate distribution of (B and D) TYRP1 immunofluoresence as revealed by staining of MEL5. Melan-a cells transfected with a Rab38-GFP expression construct. (E) Bright-field image of melanosome cell periphery and (F) matching GFP fluorescence image for transfected cell. (E) Large white arrow indicates a nontransfected cell. (E and F) Small arrows demonstrate colocalization of Rab38-GFP signal with highly pigmented end stage melanosomes at cell periphery. (Scale bar: A–D = 1.6 μM; E and F = 2.4 μM.)
Rab38cht Does Not Result in Platelet Storage Defects.
Subsets of mouse coat color mutants with mutations in genes involved in vesicular trafficking like pale ear and beige also cause platelet aggregation defects, modeling HPS and CHS, respectively. To analyze further the pathology of Rab38cht/Rab38cht mice, bleeding times were measured to assay platelet function. No difference was observed between wild-type and Rab38cht/Rab38cht mice (2.53 vs. 2.41 min; P = 0.7); this finding is consistent with Rab38cht/Rab38cht being a genocopy for the OCAIII (Tyrp1b/Tyrp1b) mouse model, but not for HPS or CHS. These data indicate that RAB38 is required for efficient targeting of TYRP1 to pigmented melanosomes and suggest that Rab38cht/Rab38cht is a genocopy of the Tyrp1b/Tyrp1b OCAIII mouse model.
Discussion
Using cDNA microarray expression profiling we have identified RAB38 as an important gene involved in melanocyte function. Analysis of microarray expression patterns demonstrated that RAB38 had an expression profile similar to nine previously identified melanocyte genes known to function in a melanocyte-specific fashion. These melanocyte genes include DCT, TRYP1, and PMEL17, which are essential for melanosome function; MELAN-A/MART1 (24) and MLSN (25), which are important melanoma antigens; AIM-1 recently identified as the gene responsible for B in medaka (26), underwhite in mouse (3), and OCA4 in human (3); CHS1, which functions in melanosome/lysosome vesicle trafficking (27); and PAX3, a paired box transcription factor that regulates melanocyte gene expression (28–30), including expression of TYRP1 (31). Mutations in seven of these genes have been identified in human and/or murine disorders associated with pigmentation variations (Fig. 1).
Rab38 was assessed as a candidate gene for the cht locus for three reasons. First, comparative genomic analysis predicted a colocalization of conserved synteny between human RAB38 and the region of the cht locus in the mouse genome. Second, expression of Rab38 was found to be restricted to those cell types affected in cht/cht mice (18) (Fig. 2). Finally, Rab38 is a member of a family of proteins known to play a crucial role in vesicular trafficking (32, 33).
Sequence analysis of the Rab38 coding region from cht mice revealed a G146T transversion in exon 1. This sequence alteration is likely the causative mutation because this allele arose as a spontaneous mutation on a C57BL/6J background. Moreover, the G146T alteration results in a Gly to Val substitution within the GTP binding pocket of RAB38. Crystal structure analysis of RAB3A, used as a model for Rab proteins, predicts that this amino acid residue directly contacts GTP in the nucleotide binding pocket (34). Furthermore, a mutation of the analogous amino acid residue in RAB5, a Rab that regulates homotypic fusion of endosomes, results in an increased rate of GDP dissociation in vitro and stimulation of endosome fusion in vivo (35). Additional support for the functional relevance of this mutation comes from studies of Ras protein. Substitutions in Ras at the analogous G13 residue, including the same G to V mutation as in Rab38cht, have been identified in acute myeloid leukemia (36, 37). Based on these observations, we predict that the G to V mutation in RAB38 would disrupt its function in vivo. Further analysis is needed to determine mode of action for this allele.
The coat color of Rab38cht/Rab38cht mice closely resembles that of the brown (Tyrp1b/Tyrp1b), OCAIII mouse model. The brown mouse model contains a defect in a melanin biosynthesis gene Tyrp1, resulting in a coat color change of the C57BL/6J mouse from black to brown. TYRP1 is a melanosomal membrane glycoprotein, which functions both as a DHICA oxidase enzyme and a provider of structural stability to TYR in the melanogenic enzyme complex. TYRP1 is believed to transit from the trans-Golgi network to stage II melanosomes by means of clathrin-coated vesicles, possibly by first passing through an uncharacterized sorting compartment (4). Based on the similar coat phenotype and predicted Rab protein function, we hypothesized that RAB38 may be specifically involved in trafficking of melanosomal proteins like TYRP1 to the melanosome. Consistent with this idea, GFP-tagged RAB38 colocalizes with melanosomes in pigmented melanocyte lines in culture, and TYRP1 is inefficiently targeted to pigmented end-stage melanosomes in Rab38cht/Rab38cht melanocytes. Thus, the brown coat color observed in Rab38cht/Rab38cht mice is predicted to result from reduced amount of melanosomal TYRP1 and implicates RAB38 in the vesicle trafficking required for proper targeting of proteins such as TYRP1 to melanosomes.
The formation of melanosomes and melanin pigment deposition within them requires a series of specific vesicular trafficking steps (2, 4). Comparison of the phenotype of Rab38cht/Rab38cht mice to other mouse mutants where defects in the trafficking of proteins has been identified should provide insight into the site of action of RAB38. Four genes involved in HPS, (HPS1, AP3β1, HPS3, and HPS4), when mutated, result in the mouse models pale ear (ep), mocha, cocoa, and light ear, respectively. For each of these mouse models the color of melanin produced by the melanosome is lighter in color or of a brown hue. Interestingly, similar to what is seen in Rab38cht/Rab38cht-derived melanocytes, melanocytes from Hps1ep/Hps1ep mutants also exhibit a mislocalization of TYRP1 into membranous complexes rather than premelanosomes (38), again yielding a brown mouse. In addition to melanosome pigment defects, HPS mice exhibit enlargement of melanosomes and lysosomes and reduced platelet aggregation (27, 39). This finding suggests involvement of HPS genes in early vesicle sorting events that affect both lysosomes as well as melanosomes. However, it appears that Rab38cht is not in the same class of mutants as those of the HPS mouse models, because Rab38cht/Rab38cht mice do not exhibit enlarged melanosomes or defects in platelet function. Thus, although both HPS1 and RAB38 appear to be involved in proper sorting of TYRP1, this regulation appears to occur at different steps in the trafficking process. Because RAB38 appears to affect only melanosome trafficking, we suggest that RAB38 is involved in vesicle trafficking downstream of HPS genes.
Rab38cht mice appear to be a genocopy of the TYRP1b, OCA mouse model, because of the essential role of RAB38 in proper TYRP1 trafficking to late-stage melanosomes, mimicking the cellular and clinical phenotype. OCA is a heterogeneous genetic disorder that has been associated with mutations in TYR (OCAI), P (OCAII), TYRP1 (OCAIII), and AIM1 (OCAIV). However, 10% of patients clinically diagnosed with OCA do not have mutations in any of these genes (Richard King and William Oetting, personal communication). Given the heterogeneity of OCA and the predicted role of RAB38 in TYRP1 sorting, we propose RAB38 as a candidate gene for patients with OCA where a molecular defect in TYR, P, TYRP1 or AIM1 has not been identified.
Acknowledgments
We acknowledge the following individuals for providing reagents: Javed Khan (National Institutes of Health) for the RH, CTR, and RHD rhabdomyosarcoma RNA; Dorothy C. Bennett for melan-a cells (St. George's Hospital, London); Belinda Harris (Jackson Laboratories) for Rab38cht/Rab38cht mice; and Piero Carninci and Yoshihide Hayashizaki (RIKEN, Genome Sciences Center, Genome Exploration Group, Yokohama, Japan) for mouse Rab38 and Aim-1 cDNA clones. We also thank Heinz Arnheiter, Vince Hearing, Joyce Dunn, Ramin Mollaaghababa, and Jill Denny for careful reading of this manuscript.
Abbreviations
- OCA
oculocutaneous albinism
- CHS
Chediak–Higashi syndrome
- HPS
Hermansky-Pudlak syndrome
- cht
chocolate
- GFP
green fluorescent protein
- RPE
retinal pigmented epithelium
- TYRP1
tyrosinase-related protein 1
Footnotes
References
- 1.Jackson I J. Hum Mol Genet. 1997;6:1613–1624. doi: 10.1093/hmg/6.10.1613. [DOI] [PubMed] [Google Scholar]
- 2.King R A, Hearing V J, Creel D, Oetting W S. In: The Metabolic Basis of Inherited Disease. 7th Ed. Scriver C R, Beaudet A L, Sly W S, Valle D V, editors. New York: McGraw–Hill; 1995. pp. 4353–4392. [Google Scholar]
- 3.Newton J M, Cohen-Barak O, Hagiwara N, Gardner J M, Davisson M T, King R A, Brilliant M H. Am J Hum Genet. 2001;69:981–988. doi: 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marks M S, Seabra M C. Nat Rev Mol Cell Biol. 2001;2:738–748. doi: 10.1038/35096009. [DOI] [PubMed] [Google Scholar]
- 5.Loftus S K, Pavan W J. Pigment Cell Res. 2000;13:141–146. doi: 10.1034/j.1600-0749.2000.130304.x. [DOI] [PubMed] [Google Scholar]
- 6.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 8.Mody M, Cao Y, Cui Z, Tay K Y, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien J Z. Proc Natl Acad Sci USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Nature (London) 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Rao K, Bowers M B, Copeland N G, Jenkins N A, Hammer J A., 3rd J Cell Sci. 2001;114:1091–1100. doi: 10.1242/jcs.114.6.1091. [DOI] [PubMed] [Google Scholar]
- 11.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 12.Loftus S K, Chen Y, Gooden G, Ryan J F, Birznieks G, Hilliard M, Baxevanis A D, Bittner M, Meltzer P, Trent J, Pavan W. Proc Natl Acad Sci USA. 1999;96:9277–9280. doi: 10.1073/pnas.96.16.9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steel K P, Davidson D R, Jackson I J. Development (Cambridge, UK) 1992;115:1111–1119. doi: 10.1242/dev.115.4.1111. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson D G, Nieto M A. In: Methods in Enzymology: Guide to Techniques in Mouse Development. Wassarman P M, DePamphilis M L, editors. San Diego: Academic; 1993. pp. 361–373. [Google Scholar]
- 15.Wu X, Bowers B, Wei Q, Kocher B, Hammer J A., 3rd J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- 16.Sviderskaya E V, Novak E K, Swank R T, Bennett D C. Genetics. 1998;148:381–390. doi: 10.1093/genetics/148.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trent J M, Stanbridge E J, McBride H L, Meese E U, Casey G, Araujo D E, Witkowski C M, Nagle R B. Science. 1990;247:568–571. doi: 10.1126/science.2300817. [DOI] [PubMed] [Google Scholar]
- 18.Jager D, Stockert E, Jager E, Gure A O, Scanlan M J, Knuth A, Old L J, Chen Y T. Cancer Res. 2000;60:3584–3591. [PubMed] [Google Scholar]
- 19.Potter M D, Rinchik E M. Mamm Genome. 1993;4:46–48. doi: 10.1007/BF00364663. [DOI] [PubMed] [Google Scholar]
- 20.Macpike A, Mobraaten L E. Mouse News Lett. 1984;700:86. [Google Scholar]
- 21.Hearing V J, Phillips P, Lutzner M A. J Ultrastruct Res. 1973;43:88–106. doi: 10.1016/s0022-5320(73)90072-5. [DOI] [PubMed] [Google Scholar]
- 22.Schimmoller F, Simon I, Pfeffer S R. J Biol Chem. 1998;273:22161–22164. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- 23.Chavrier P, Goud B. Curr Opin Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y T, Stockert E, Jungbluth A, Tsang S, Coplan K A, Scanlan M J, Old L J. Proc Natl Acad Sci USA. 1996;93:5915–5919. doi: 10.1073/pnas.93.12.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan L M, Deeds J, Cronin F E, Donovan M, Sober A J, Kauffman M, McCarthy J J. J Clin Oncol. 2001;19:568–576. doi: 10.1200/JCO.2001.19.2.568. [DOI] [PubMed] [Google Scholar]
- 26.Fukamachi S, Shimada A, Shima A. Nat Genet. 2001;28:381–385. doi: 10.1038/ng584. [DOI] [PubMed] [Google Scholar]
- 27.Introne W, Boissy R E, Gahl W A. Mol Genet Metab. 1999;68:283–303. doi: 10.1006/mgme.1999.2927. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe A, Takeda K, Ploplis B, Tachibana M. Nat Genet. 1998;18:283–286. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- 29.Potterf S B, Furumura M, Dunn K J, Arnheiter H, Pavan W J. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 30.Hornyak T J, Hayes D J, Chiu L Y, Ziff E B. Mech Dev. 2001;101:47–59. doi: 10.1016/s0925-4773(00)00569-4. [DOI] [PubMed] [Google Scholar]
- 31.Galibert M D, Yavuzer U, Dexter T J, Goding C R. J Biol Chem. 1999;274:26894–26900. doi: 10.1074/jbc.274.38.26894. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen E, Severin F, Backer J M, Hyman A A, Zerial M. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 33.Scott G, Zhao Q. J Invest Dermatol. 2001;116:296–304. doi: 10.1046/j.1523-1747.2001.01221.x. [DOI] [PubMed] [Google Scholar]
- 34.Dumas J J, Zhu Z, Connolly J L, Lambright D G. Struct Fold Des. 1999;7:413–423. doi: 10.1016/s0969-2126(99)80054-9. [DOI] [PubMed] [Google Scholar]
- 35.Li G, Liang Z. Biochem J. 2001;355:681–689. doi: 10.1042/bj3550681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bos J L, Toksoz D, Marshall C J, Verlaan-de Vries M, Veeneman G H, van der Eb A J, van Boom J H, Janssen J W, Steenvoorden A C. Nature (London) 1985;315:726–730. doi: 10.1038/315726a0. [DOI] [PubMed] [Google Scholar]
- 37.Stirewalt D L, Kopecky K J, Meshinchi S, Appelbaum F R, Slovak M L, Willman C L, Radich J P. Blood. 2001;97:3589–3595. doi: 10.1182/blood.v97.11.3589. [DOI] [PubMed] [Google Scholar]
- 38.Sarangarajan R, Budev A, Zhao Y, Gahl W A, Boissy R E. J Invest Dermatol. 2001;117:641–646. doi: 10.1046/j.0022-202x.2001.01435.x. [DOI] [PubMed] [Google Scholar]
- 39.Swank R T, Novak E K, McGarry M P, Zhang Y, Li W, Zhang Q, Feng L. Pigment Cell Res. 2000;13:59–67. doi: 10.1034/j.1600-0749.13.s8.12.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Addess K J, Geer L, Madej T, Marchler-Bauer A, Zimmerman D, Bryant S H. Nucleic Acids Res. 2000;28:243–245. doi: 10.1093/nar/28.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 42.Ostermeier C, Brunger A T. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]