Figure 1.
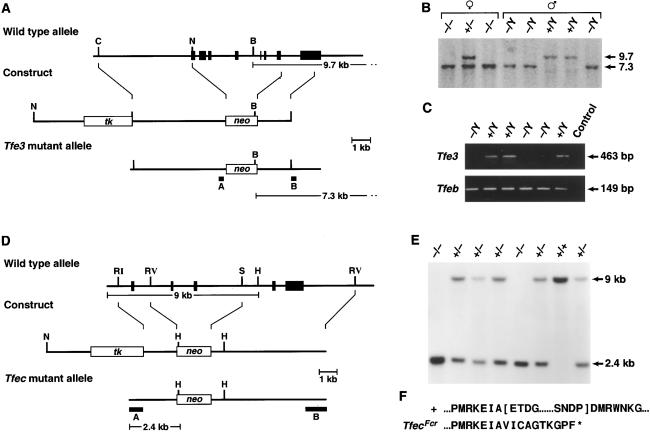
Generation of Tfe3Fcr and TfecFcr mutant mice. (A) The Tfe3 knockout. The exon-intron organization of Tfe3 is shown at the top with exons indicated as black boxes. A 5.5-kb genomic fragment containing seven Tfe3 exons, the 5′ end of Tfe3, and the bHLH-Zip domains was replaced with PgkNeo. The first exon corresponds to nucleotides 14–153 of mouse Tfe3 (19). (B) Identification of Tfe3Fcr mutant animals. Tail DNA from the progeny of a heterozygous Tfe3Fcr intercross was digested with BamHI and analyzed by Southern analysis with probe B, which detects 9.7- (wild-type) and 7.3-kb (mutant) alleles. Tfe3Fcr males only transmit the mutant allele to their daughters, confirming the X chromosome linkage of Tfe3 (19). (C) Tfe3 expression in mutant animals. Kidney RNA from Tfe3Fcr and wild-type males was reversed-transcribed and PCR-amplified by using Tfe3 primers 5′-ccaagctggcttcccaggctctcac and 5′-gttaatgttgaatcgcctgcgtcg. The wild-type 463-bp fragment corresponds to nucleotides 245–708 of Tfe3 (19). The same samples were also amplified with Tfeb primers 5′-ccctgtctagcagccacctgaacg and 5′-gccgctccttggccagggctctgctc as a control. Primer pairs spanned exon-intron boundaries to eliminate false positives. (D) The Tfec knockout. A 5-kb fragment containing two Tfec bHLH exons was replaced with PgkNeo. Deleted exons correspond to nucleotides 507–639 of rat Tfec (18). (E) Identification of TfecFcr mutant animals. Tail DNA from the progeny of a heterozygous mutant intercross was digested with HindIII and analyzed by Southern analysis with probe A, which detects 9.0- (wild-type) and 2.4-kb (mutant) alleles. (F) Sequence of the TfecFcr mutant transcript. Kidney RNA from wild-type and mutant animals was reverse-transcribed and PCR-amplified with Tfec primers 5′-cagtgatgctggctgtgc and 5′-gtagccacttgatgtagtcc. Sequencing of the mutant transcript showed that it lacked two conserved Tfec functional domains (indicated by brackets in the wild-type sequence) and is out of frame for the rest of the coding region. B, BamHI; C, ClaI; N, H, HindIII; RI, EcoRI; N, NotI; RV, EcoRV; S, SacI.