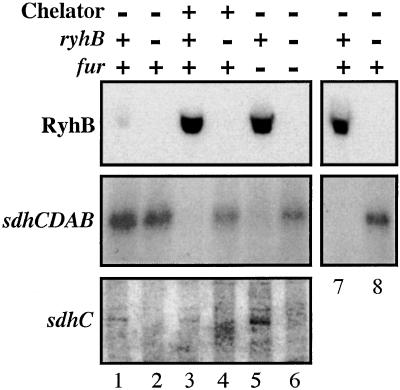
Figure 3.
Cells were grown in LB (RyhB and sdhCDAB, lanes 1–6) or M63 glycerol (RyhB and sdhCDAB , lanes 7 and 8; and sdhC, lanes 1–6) to an OD600 0.3 and 2,2′-dipyridyl was added to a final concentration of 250 μM where indicated. A sample was removed 15′ after chelator addition; RNA was isolated as described in Materials and Methods. For lanes 1, 3, and 7, a fur+ ryhB+ strain (EM1055) was used; for lanes 2, 4, and 8, a fur+ ΔryhB1∷cat strain, EM1238, was used. Cells for lanes 5 (fur− ryhB+) and 6 (fur− ΔryhB1∷cat) were strains EM1256 and EM1257, respectively. [Top (RyhB)] For determination of RyhB amount, 3 μg of total RNA samples extracted from cells grown in LB (lanes 1–6) or M63 glycerol media (lanes 7 and 8) were loaded on an 8% PAGE gel. After migration, a Northern blot analysis was performed with oligoprobe EM1 (Table 1). [Middle (sdhCDAB)] Ten micrograms of the same total RNA extracts as in Top were loaded on a denaturating 1.2% agarose gel. After migration, a Northern blot hybridization was performed with a specific oligoprobe for sdhC (probe EM8 shown in Fig. 2, Table 1). [Bottom (sdhC)] Three micrograms of RNA extracted from cells grown in M63 glycerol media were loaded on a 4% PAGE gel. A Northern blot was performed by using oligonucleotide probe EM8 within sdhC.