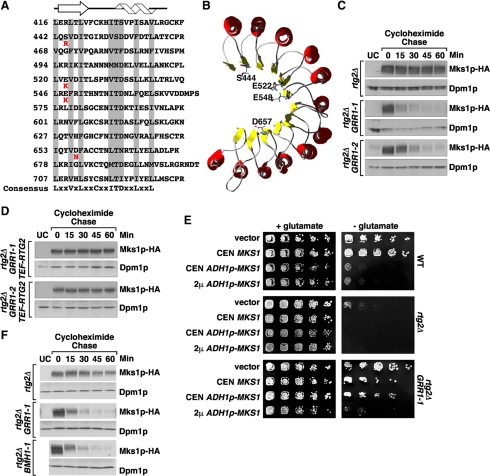
Figure 2.
GRR1 dominant mutations destabilize Mks1p in rtg2Δ cells. (A) Sequence alignment of the Grr1p leucine-rich repeats (LRR). The conserved residues are highlighted in gray and the consensus sequence shown at the bottom indicates amino acid residues present in at least five of the repeats. The altered residue of each of the four dominant mutations of GRR1-(1-4) is indicated in red underneath the wild-type residue. The predicted secondary structure (helix for α-helix and arrow for β-sheet) is indicated at the top. (B) The dominant GRR1 mutations localize to the concave surface of the LRR of Grr1p. The modeled 3D structure is adapted from Hsiung et al. (2001). The residues mutated in the dominant GRR1 mutants are indicated by showing their side chains. (C) Mks1p expressed from the ADH1 promoter is unstable in rtg2Δ GRR1-1 and rtg2Δ GRR1-2 cells. Mks1p stability was examined in indicated cells using cycloheximide chase assay as described in Materials and Methods.. Mks1p-HA levels were determined by probing Western blots with anti-HA antibody. Dpm1p, dolichol phosphate mannose synthase, was used as loading control. UC, untagged control. (D) Rtg2p protects Mks1p from degradation in GRR1-1 and GRR1-2 mutants. Mks1p stability was examined following a cycloheximide chase in rtg2Δ GRR1-1 and rtg2Δ GRR1-2 cells, each containing a copy of RTG2 integrated at the LEU2 locus under the control of the TEF1 promoter. (E) Overexpression of MKS1 reverses the rtg2Δ bypass phenotype of the GRR1-1 mutation. Serial dilutions of culture of indicated cells were prepared and cells were spotted onto solid YNBD medium with or without 0.02% glutamate. Cells were grown for three days at 30°C. (F) Stability assay of Mks1p-HA expressed from its own promoter. Mks1p stability was examined using cycloheximide chase assay.