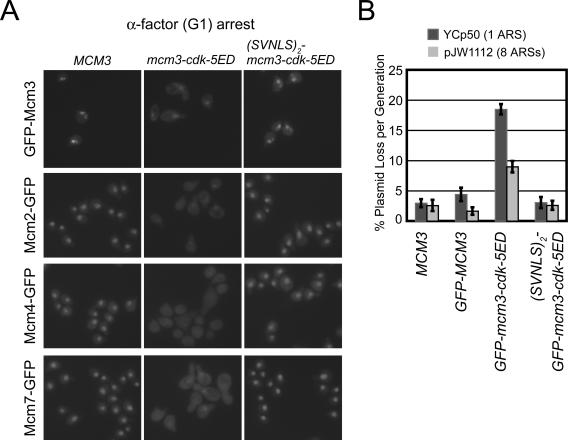
Figure 10.
Phosphomimic mutations of the Mcm3 CDK consensus sites promote net nuclear export of Mcm proteins in G1 phase and this mislocalization impairs replication initiation. (A) (First Row) YJL2160 (GFP-MCM3), YJL1265 (GFP-mcm3-cdk5ED), YJL1260 (SVNLS2-GFP-mcm3-cdk5ED). (Second Row) YJL2033 (mcm2::{MCM2-GFP}), YJL4162 (mcm2::{MCM2-GFP} mcm3-cdk5ED), YJL4094 (mcm2::{MCM2-GFP} SVNLS2-mcm3-cdk5ED). (Third Row) YJL2037 (mcm4::{MCM4-GFP}), YJL4103 (mcm4::{MCM4-GFP} mcm3-cdk5ED), YJL4098 (mcm4::{MCM4-GFP} SVNLS2-mcm3-cdk5ED). (Fourth Row) YJL2217 (mcm7::{MCM7-GFP}), YJL4108 (mcm7::{MCM7-GFP} mcm3-cdk5ED), YJL4096 (mcm7::{MCM7-GFP} SVNLS2-mcm3-cdk5ED). All strains were arrested in G1 phase with α-factor for 90 min (>95% unbudded) before being examined by fluorescence microscopy. (B) Mcm mislocalization due to the mcm3-cdk5ED mutation impairs replication initiation. Plasmid loss rates per were measured over 12-20 generations for plasmids YCp50 (1 ARS) and pJW1112 (8 ARSs) in YJL310 (MCM3), YJL2160 (GFP-MCM3), YJL1265 (GFP-mcm3-cdk5ED), and YJL1259 (SVNLS2-GFP-mcm3-cdk5ED). Histogram shows average loss rate per generation and SE for four independent isolates of each plasmid-yeast pair. Complete failure to replicate a plasmid would result in a theoretical loss rate of 50% per generation.