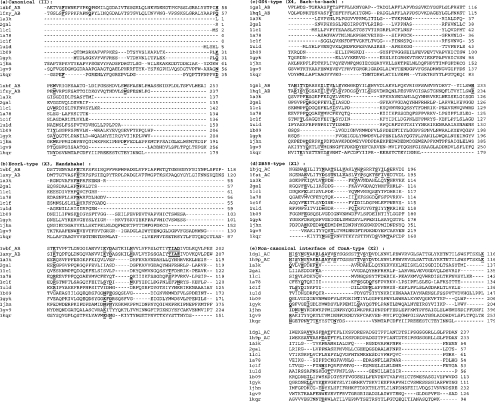
Figure 3. Multiple sequence alignments of different legume lectins with galectins, pentraxins, calnexin, calreticulin and Vp4 sialic-acid-binding domain.
Only a few representatives of each type, and the sequence fragments involving the signature motifs, are shown. Types II, X1, X2, X3 and X4 interface types of the legume lectins are shown. The first two sequences in each type belong to the characteristic legume lectins of the particular type. The residues forming the signature sequence motifs of each legume lectin interface type are highlighted in bold and underlined in the first two legume lectin sequences of each interface type. The other lectins included in the alignments are, galectin-1 (1a78), congerin [conger-eel (Conger conger) galectin; 1c1f], galectin-2 (1uld), galectin-3 (1a3k), Charcot–Leyden protein (1lcl), galectin-7 (2gal), human CRP (pentraxin, 1b09), human SAP (pentraxin, 1gyk), calnexin (1jhn), calreticulin (1gv9) and Vp4 sialic-acid-binding domain (1kqr). The residues in these lectins, which are conserved or conservatively mutated at the signature motif positions of legume lectins, are also shown in bold and underlined. The residue numbers are indicated at the end of each line. It can be seen from these alignments that although some residues from the signature motifs are conserved in the galectins, pentraxins, calnexin, calreticulin and Vp4 sialic-acid-binding domain, most of the residues required for any of the legume lectin interfaces are absent, thereby excluding these legume lectin interface types in these lectins.