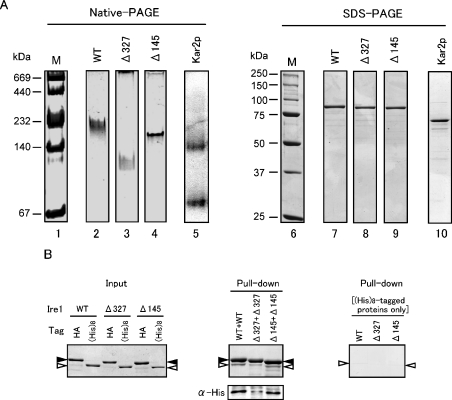
Figure 5. Dimer-forming ability of core-stress sensing region in vitro.
(A) Native PAGE analysis of the core-stress sensing region. Native PAGE (4–20% gradient gel, left-hand panel) or SDS/PAGE (10% gel, right-hand panel) of MBP–rP[II-IV]–His8 (marked WT) and its Δ327 and Δ145 mutant versions was performed as described in the Experimental section. The gel was stained with Coomassie Brilliant Blue except for lane 5 (immunoblotting with anti-Kar2 antibody) and photographed. Lanes 1 and 6, the High Molecular Weight Native Marker Kit (Amersham Biosciences) and Precision Protein standards (Bio-Rad) respectively, as molecular-mass markers (M) (sizes are indicated in kDa); lanes 2 and 7, MBP–rP[II-IV]–His8; lanes 3 and 8, Δ327 mutant; lanes 4 and 9, Δ145 mutant; lanes 5 and 10, yeast BiP protein. (B) In vitro pull-down assay to demonstrate the self-binding ability of the core stress-sensing region. Left-hand panel: 0.625 μg of the HA-tagged proteins {MBP–rP[II-IV]–HA (WT), and its Δ327 or Δ145 mutant} and 0.5 μg of the His8-tagged proteins {MBP–rP[II-IV]–His8 (WT), and its Δ327 or Δ145 mutant} were subjected to SDS/6% PAGE, and the gel was stained with Coomassie Blue. Positions of the HA-tagged and His8-tagged proteins were respectively marked by black and white arrowheads. Middle panel: 5 μg of the HA-tagged proteins and 4 μg of the His8-tagged proteins were subjected to the pull-down assay using anti-HA antibody–agarose beads as described in the Experimental section, and total (upper panel) or a 1/6 (lower panel) volume of the bead-bound proteins were fractionated by SDS/6% PAGE. Then the gels were stained with Coomassie Blue (upper panel) or subjected to anti-His tag Western blotting (lower panel). Positions of the HA-tagged and His8-tagged proteins were marked as in the left-hand panel. Right-hand panel: a similar experiment was performed as that shown in the middle panel, except for the absence of the HA-tagged proteins.