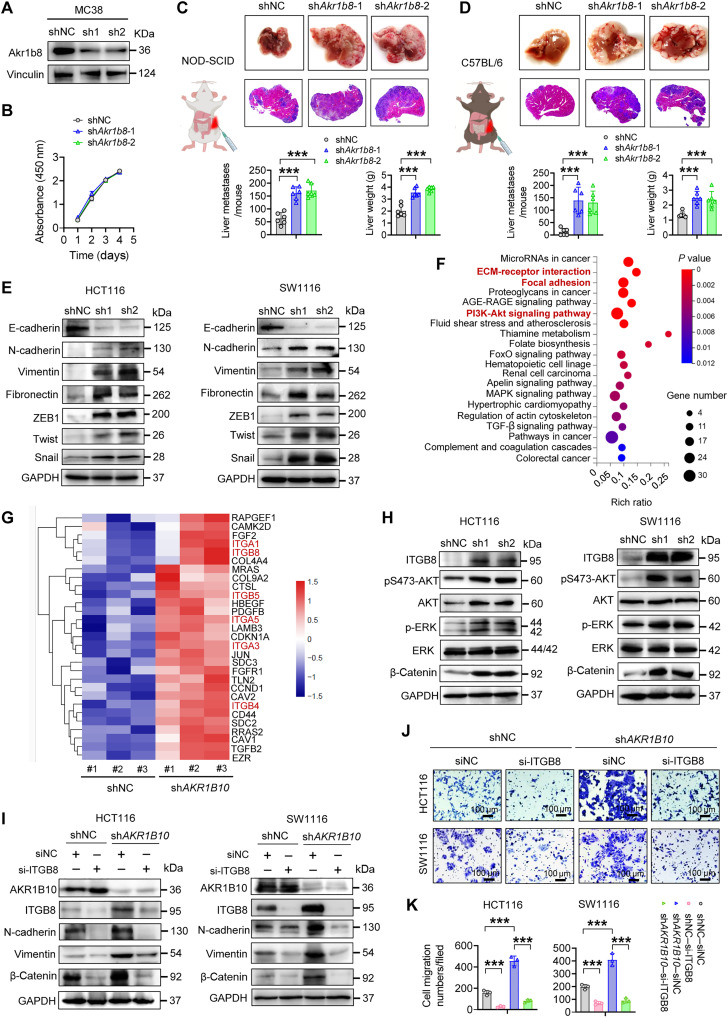
Fig. 3. AKR1B10 suppresses EMT by down-regulating integrin signaling.
(A) Western blot analysis of Akr1b8 knockdown efficiency in MC38 cells. (B) Cell proliferation rates of MC38 (shNC and shAkr1b8) cells assessed using the CCK-8 assay (n = 4). (C) Representative images of liver metastatic nodules in NOD-SCID mice on day 15 following splenic injection of MC38 shNC and shAkr1b8 cells. Liver weight and the number of metastases were quantified (n = 6 mice per group). (D) Representative images of liver metastatic nodules in C57BL/6 mice on day 18 following splenic injection of MC38 (shNC and shAkr1b8) cells. Liver weight and the number of metastases were quantified (n = 6 mice per group). (E) Western blot analysis of EMT markers and EMT-related TFs in HCT116 and SW1116 cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (F) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the top 20 up-regulated biological processes in shAKR1B10 cells based on RNA sequencing (RNA-seq). PI3K, phosphatidylinositol 3-kinase; MAPK, mitogen-activated protein kinase; TGF-β, transforming growth factor–β; AGE-RAGE, Advanced Glycation End products–Receptor for Advanced Glycation End products; FoxO, Forkhead box O. (G) Heatmap depicting mRNA levels of genes from selected KEGG pathways. (H) Western blot analysis of ITGB8 protein and its downstream factors in HCT116 and SW1116 cells. p-ERK, phosphorylated ERK. (I) Western blot analysis of ITGB8 protein and its downstream EMT markers in HCT116 and SW1116 cells after ITGB8 silencing. (J and K) Representative images (J) and quantification (K) of transwell assays in HCT116 and SW1116 cells after ITGB8 silencing. (C, D, and K) n = 3. Data are presented as mean ± SD, with two-way ANOVA test (B) or unpaired Student’s t test (C, D, and K). ***P < 0.001.