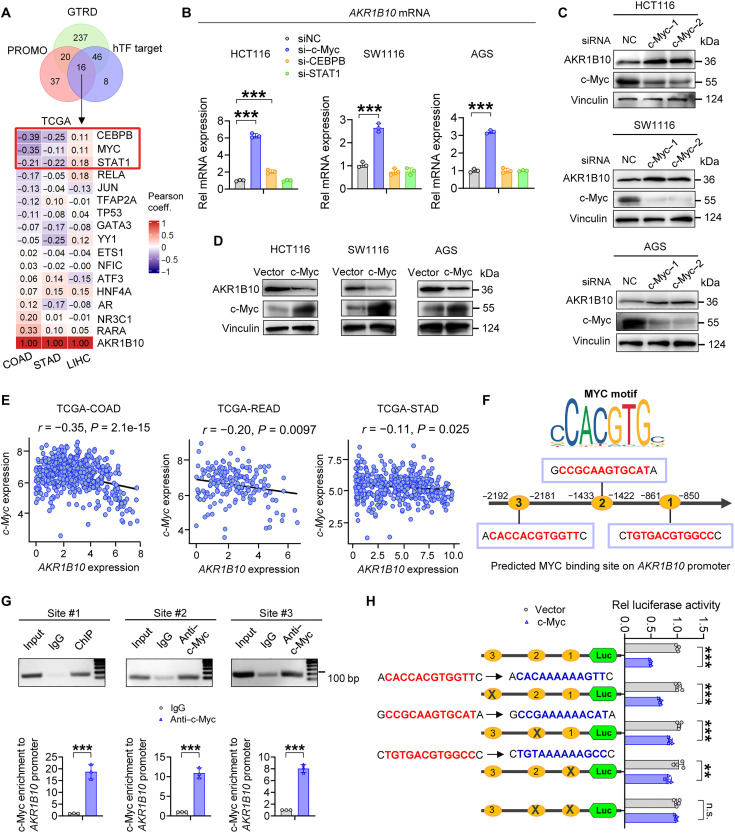
Fig. 8. c-Myc suppresses AKR1B10 transcription in gastrointestinal cancers.
(A) Bioinformatics analysis identifying three putative TFs that regulate AKR1B10 expression and show an inverse coexpression pattern with AKR1B10 between gastrointestinal cancer and HCC, based on TCGA database. coeff., coefficient. (B) Quantitative reverse transcription (qRT)–PCR analysis of AKR1B10 mRNA expression in HCT116, SW1116, and AGS cells transfected with small interfering RNAs (siRNAs) targeting c-Myc, CEBPB, or STAT1. (C and D) Western blot analysis of AKR1B10 protein expression in HCT116, SW1116, and AGS cells after c-Myc knockdown (C) or overexpression (D). (E) Correlation between AKR1B10 and c-Myc mRNA levels in human COAD (n = 494), READ (n = 173), and stomach adenocarcinoma (STAD; n = 405) tissues, based on TCGA database. (F) Identification of putative c-Myc binding sites within the AKR1B10 gene promoter region using the JASPAR database. (G) ChIP-qPCR analysis depicting the binding sites of c-Myc within the AKR1B10 promoter region in HCT116 cells. (H) Dual-luciferase reporter assay showing that mutations at both site 1 (−850 ~ −861) and site 2 (−1422 ~ −1433) abrogated c-Myc–mediated repression of AKR1B10 promoter activity. Luc, luciferase. Data in (B), (G), and (H) are presented as mean ± SD (n = 3), with unpaired Student’s t test. ***P < 0.001.