Abstract
Protein misfolding is a contributor to the development of type 2 diabetes (T2D), but the specific role of impaired proteostasis is unclear. Here we show a robust accumulation of misfolded proteins in the mitochondria of human pancreatic islets from patients with T2D and elucidate its impact on β cell viability through the mitochondrial matrix protease LONP1. Quantitative proteomics studies of protein aggregates reveal that islets from donors with T2D have a signature resembling mitochondrial rather than endoplasmic reticulum protein misfolding. Loss of LONP1, a vital component of the mitochondrial proteostatic machinery, with reduced expression in the β cells of donors with T2D, yields mitochondrial protein misfolding and reduced respiratory function, leading to β cell apoptosis and hyperglycaemia. LONP1 gain of function ameliorates mitochondrial protein misfolding and restores human β cell survival after glucolipotoxicity via a protease-independent effect requiring LONP1-mitochondrial HSP70 chaperone activity. Thus, LONP1 promotes β cell survival and prevents hyperglycaemia by facilitating mitochondrial protein folding. These observations provide insights into the nature of proteotoxicity that promotes β cell loss during the pathogenesis of T2D, which could be considered as future therapeutic targets.
Subject terms: Diabetes, Protein aggregation, Metabolism, Proteomics, Type 2 diabetes
LONP1, whose expression is downregulated in islets from donors with type 2 diabetes, is vital to mediate efficient mitochondrial protein folding, thus preventing proteotoxicity and promoting islet β cell survival and function.
Main
Optimal protein homeostasis, also known as proteostasis, is vital to combating ageing-related diseases, including cancer, cardiovascular disorders, neurodegenerative diseases and type 2 diabetes (T2D)1. Impairments in proteostasis include a decline in correct protein folding, assembly and turnover, and coordinated balance of protein stoichiometry required to form multi-subunit complexes2. Emerging evidence demonstrates that T2D is a protein misfolding-related disease3. Among the best examples of protein misfolding in T2D occur in pancreatic β cells, where misfolding of proinsulin and islet amyloid polypeptide (IAPP), a hormone co-secreted with insulin, lead to β cell dysfunction4,5. Proinsulin misfolding precipitates endoplasmic reticulum (ER) stress, while aggregates of IAPP lead to a cascade of defects culminating in β cell apoptosis4,5. Accordingly, ER protein misfolding and stress are often considered major mediators of β cell apoptosis in T2D6,7. However, beyond proinsulin and IAPP, the extent, location and specific impact of protein misfolding in T2D is unclear.
A parallel and related feature of the importance of disrupted proteostasis to ageing-related diseases and T2D is the development of mitochondrial dysfunction. Mitochondria are crucial for several vital functions in pancreatic β cells, including the support of fuel-stimulated insulin release and maintenance of β cell mass and survival8–10. Indeed, β cells from islet donors with T2D develop dilated mitochondrial ultrastructure with dysmorphic cristae, as well as bioenergetic defects11,12. The observation of increases in reactive oxygen species (ROS) and β cell oxidative damage in individuals with T2D may also be closely related to mitochondrial damage because mitochondria are a major source of ROS production13. Furthermore, recent work identified that β cell mitochondrial gene expression and oxidative phosphorylation (OXPHOS) defects precede the development of T2D14. Human genetic studies also support associations between mitochondria and T2D15–21. However, a mechanistic link between impairments in β cell proteostasis and mitochondrial health in T2D has not been examined.
Most of the mitochondrial proteome is encoded by the nuclear genome and synthesized on cytosolic ribosomes. Optimal mitochondrial proteostasis depends on the correct import and folding of unfolded mitochondrial precursors into their functional structures, as well as safeguard mechanisms to respond to protein misfolding or stress. Within the mitochondrial matrix, chaperone complexes consisting of mitochondrial HSP70 (mtHSP70) (also known as HSPA9, GRP75 and mortalin), DNAJA3 and GRPEL1 and GRPEL2, as well as mtHSP60 and mtHSP10, promote protein folding upon import22,23. Mitochondrial chaperones can also be mobilized under stress or in response to mitochondrial protein aggregates22. Indeed, exposure of islets to the saturated fatty acid palmitate to elicit lipotoxicity upregulates mtHSP70 expression24. Mitochondrial proteases not only degrade misfolded or damaged mitochondrial proteins but also have regulatory functions beyond protein clearance, including in the maintenance of the electron transport chain (ETC) and mtDNA22. Several well-known mitochondrial matrix proteases, such as LONP1 and CLPXP, govern mitochondrial protein quality control; mutations in mitochondrial proteases have been linked to neurodegenerative diseases, cancer and eye diseases25–27. LONP1 is a multifunctional mitochondrial AAA+ matrix protease, which turns over misfolded mitochondrial proteins, remodels the ETC–OXPHOS system during tumorigenesis and possesses chaperone-like activity by partnering together with mtHSP70 (refs. 28–30). Furthermore, LONP1 is upregulated in human islets after lipotoxicity31. However, the importance of LONP1’s action and its partnership with mtHSP70 in β cells have not yet been explored.
In this study, we demonstrate a crucial link between mitochondrial protein misfolding in T2D and the development of β cell failure. Using quantitative proteomics in human islets, genetic mouse models, high-resolution imaging and biochemical assays, we elucidate that impairments in mitochondrial protein folding, which we observed in the human islets of donors with T2D, elicit β cell apoptosis, ultimately leading to loss of β cell mass and hyperglycaemia. Proteomics studies revealed that insoluble and aggregated proteins in human islets of donors with T2D surprisingly more closely resembled mitochondrial rather than ER protein misfolding. Importantly, loss of LONP1, whose expression is reduced in the β cells of donors with T2D, induces mitochondrial protein misfolding, which leads to mitochondrial structural and respiratory defects, oxidative stress, DNA damage and impairments in ETC assembly. Furthermore, we demonstrate that β cell death resulting from LONP1 deficiency is driven by mitochondrial protein misfolding and not oxidative stress, as shown using genetically encoded and pharmacological antioxidants. Moreover, our results support that the chaperone-like activity of LONP1, and not the protease activity of LONP1, prevents mitochondrial protein misfolding to promote cell survival and to protect against glucolipotoxicity (GLT) in mouse and human β cells. Thus, our results illustrate the importance of LONP1-mediated mitochondrial protein folding to defend against β cell loss in T2D.
Results
Insoluble mitochondrial proteins are enriched in human islets from donors with T2D
We initially took advantage of a validated biochemical approach to evaluate protein solubility in cells or tissues because insoluble proteins will include misfolded proteins or aggregates29,32,33. Briefly, human islets isolated from donors with or without T2D were lysed with a buffer containing 1% Triton X-100 (refs. 29,32–36), a widely used and reliable non-ionic detergent for the solubilization of proteins, including lipid-soluble membrane proteins and integral membrane proteins34,35. This was followed by centrifugation to isolate detergent-soluble and detergent-insoluble protein fractions. Indeed, the Triton X-100 insoluble fraction has been previously shown to include misfolded proteins and protein aggregates29,36. We then applied unbiased quantitative proteomics approaches in both fractions using tandem mass tag (TMT) labelling and liquid chromatography–mass spectrometry (LC–MS) (Fig. 1a).
Fig. 1. Insoluble mitochondrial proteins are enriched in human islets from donors with T2D.
a, Schematic diagram illustrating the Triton X-100 approach to quantitatively examine protein solubility. b, Volcano plot of differentially expressed insoluble proteins from donors with T2D compared to Ctrls without T2D determined using −log10(P > 1.3) and log2 fold change greater than 0.1. Mitochondrial proteins (curated from MitoCarta3.0) are highlighted in green. n = 4 independent islet donors per group. c, GO cellular component analysis of significantly upregulated insoluble proteins in T2D islets. n = 4 independent islet donors per group. d, Volcano plot of differentially expressed soluble proteins in T2D islets. n = 4 independent islet donors per group. e, GO cellular component analysis of significantly downregulated soluble proteins in T2D islets. n = 4 independent islet donors per group. f, Representative immunoblot images of selected mitochondrial proteins of human islets. g, Quantification of mitochondrial insoluble proteins as fold densitometry of the insoluble/soluble protein ratio. n = 4 independent human islet donors per group. VDAC1 serves as a soluble mitochondrial protein loading control. Vinculin serves as a loading control for both soluble and insoluble fractions. h, Representative deconvolution immunofluorescence image (n = 5 for Ctrl and n = 3 for T2D) depicting mtHSP70 expression and localization from pancreatic sections of human islet donors. i,j, Volcano plots for differentially expressed proteins in the insoluble (i) and soluble (j) fractions of non-diabetic islets exposed to 1 μM CDDO or vehicle for 24 h. n = 4 independent islet donors per group. k,l, Volcano plots for differentially expressed proteins in the insoluble (k) and soluble (l) fractions of non-diabetic islets exposed to 1 μg ml−1 TUN or vehicle for 24 h. n = 4 independent islet donors per group. m, UpSet blot visualizing the intersections in insoluble protein enrichment among the T2D, CDDO and TUN groups. All data are presented as the mean ± s.e.m. b,d,i–l, P < 0.05 was determined using a two-tailed limma moderated t-test. c, P < 0.05 was determined using a hypergeometric test followed by multiple hypotheses testing using false discovery rate (FDR)-corrected P values (FDR < 0.05). g, *P < 0.05, **P < 0.01 was determined using an unpaired, two-tailed Student’s t-test. h, Scale bar, 6.25 μm. DEG, differentially expressed gene; ESCRT, endosomal sorting complexes required for transport; MS3, three-stage mass spectrometry; ND, not determined; P, insoluble fraction; S, soluble fraction.
Evaluation and application of our proteomics data as a candidate-based approach revealed 364 proteins that were differentially enriched in the insoluble fraction of islets from donors with T2D (Fig. 1b and Supplementary Data File 1). Insoluble proteins in these islets included proteins with expected functions, such as peptidyl-prolyl cis-trans isomerase C (ref. 37), which maintains ER redox homeostasis, and α-B-crystallin38, which functions as a chaperone capable of binding misfolded proteins and toxic amyloid aggregates (Fig. 1b). Importantly, Gene Ontology (GO) analysis of differentially enriched proteins in the insoluble fraction of the islets of donors with T2D revealed a high frequency of proteins localized to the mitochondria, with a striking enrichment of mitochondrial matrix proteins (Fig. 1b,c). We next overlaid insoluble proteins on MitoCarta3.0 (ref. 39), which contains a compendium of proteins with high confidence of localization to the mitochondria, and again confirmed the high frequency of mitochondrial proteins in the insoluble fraction of islets from donors with T2D (110 of 364 differentially enriched proteins; Fig. 1b, green). Many of these enriched insoluble proteins were associated with mitochondrial gene expression, protein translation and oxidative metabolism (Extended Data Fig. 1a,b). Within the soluble fraction of islets from donors with T2D, we observed a reduction of key β cell proteins, including insulin and glucokinase (Fig. 1d and Supplementary Data File 2). GO analysis of proteins differentially expressed in the soluble fraction of islets from donors with T2D included reductions in proteins associated with vesicular transport and secretion, possibly related to reductions in insulin and its secretory granules, as well as increases in RNA binding, processing and splicing, possibly related to increases in alternative splicing reported in stressed islets in diabetes (Fig. 1e and Extended Data Fig. 1c,d)40,41. Importantly, many mitochondrial proteins were lower in the soluble fraction of islets of donors with T2D and reflected proteins that were increased in the insoluble fraction, including those localized to the mitochondrial matrix and ribosome, and related to mitochondrial translation and oxidative metabolism (Fig. 1b–e and Extended Data Figs. 1a,b and 2a,b). Parallel reductions in soluble mitochondrial matrix, ribosomal and oxidative metabolism components, together with increases in these components in the insoluble fraction of the islets of donors with T2D, indicate an unexpected and remarkable shift in mitochondrial protein solubility in T2D, which could be attributed to mitochondrial protein misfolding.
Extended Data Fig. 1. Pathway analyses of quantitative proteomics of soluble and insoluble fractions from human islet donors with or without T2D.
(a) GO biological process (left) and molecular function (right) analysis of significantly upregulated insoluble proteins from human islet donors with T2D compared to non-diabetic controls. n = 4 independent islet donors/group. (b) GO biological process (left) and molecular function (right) analysis of significantly downregulated insoluble proteins from human islet donors with T2D compared to non-diabetic controls. n = 4 independent islet donors/group. (c) GO cellular component (left) and biological process (right) analysis of significantly upregulated soluble proteins from human islet donors with T2D compared to non-diabetic controls. n = 4 independent islet donors/group. (d) GO molecular function analysis of significantly upregulated soluble proteins from human islet donors with T2D compared to non-diabetic controls. n = 4 independent islet donors/group. (e) Venn diagram displaying the overlap between differentially enriched mitochondrial proteins from the insoluble fraction of human islets from donors with T2D and experimentally validated mitochondrial long-lived proteins42. (f) Quantification of protein expression of insoluble fraction (left) and soluble fraction (right) by densitometry from studies in Fig. 1f as fold change compared to control of insoluble and soluble protein expression normalized to VINCULIN. n = 4 independent human islet donors/group. *P < 0.05 by unpaired two-tailed Student’s t-test. (g) LONP1 protein densitometry (normalized to VINCULIN) in human islets only from donors with T2D and non-diabetic control donors used for TMT-MS studies. n = 4 independent human islet donors/group. Data are presented as mean ± SEM. Statistical analysis: 1A-D *P < 0.05 by hypergeometric test followed by multiple hypothesis testing using false discovery rate (FDR)-corrected P values (FDR < 0.05).
Extended Data Fig. 2. MitoPathways assessment of soluble and insoluble proteins from islet donors with T2D.
(a) Differential expression heatmap of significantly upregulated mitochondrial proteins in the insoluble fraction of T2D islets compared to non-diabetic controls. n = 4 independent islet donors/group. Proteins are categorized based on annotation from MitoPathways3.0. Black boxes separate different categories of mitochondrial proteins. (b) Differential expression heatmap of significantly downregulated mitochondrial proteins in the soluble fraction of T2D islets compared to non-diabetic controls. Proteins are categorized based on annotation from MitoPathways3.0. Black boxes used to create space to separate different categories of mitochondrial proteins for readability.
We next questioned whether the insoluble mitochondrial proteins in human islets of donors with T2D are newly synthesized and imported proteins, or long-lived mitochondrial resident proteins. A recent study revealed high longevity for a subset of mitochondrial proteins, including in the cristae subcompartment42. Mitochondrial long-lived proteins include proteins assembled into OXPHOS complexes with remarkable longevity and limited subunit exchange throughout their lifetimes, which could raise their susceptibility to aggregation. To determine whether insoluble mitochondrial proteins in human islets from donors with T2D are long-lived or short-lived, newly synthesized or imported proteins, we compared the 110 insoluble mitochondrial proteins enriched in the islets of donors with T2D with a list of 76 experimentally validated mitochondrial long-lived proteins and found that 17 of these proteins were indeed long-lived (Extended Data Fig. 1e). The observation of both insoluble short-lived and long-lived mitochondrial proteins in the islets of donors with T2D could be suggestive of a defect in mitochondrial proteostasis affecting mitochondrial resident proteins rather than solely abnormalities in the import of newly synthesized proteins.
Defects in mitochondrial proteostasis often lead to the recruitment of mitochondrial chaperones and proteases to respond to an accumulation of misfolded proteins or aggregates22. Our proteomics data led us to evaluate the presence of mitochondrial proteases and chaperones in the insoluble fraction of the human islets of donors with T2D. Indeed, we observed significant enrichment of the mitochondrial matrix proteases LONP1 and CLPX and the chaperone mtHSP70 (also known as HSPA9) in the insoluble fraction of T2D islets (Fig. 1b and Extended Data Fig. 2a). Mitochondrial proteases and chaperones are also crucial to maintain the ETC–OXPHOS machinery. Consistent with our proteomics studies, proteins in the ETC–OXPHOS system (ACO2, NDUFA10 and ATP5A), as well as LONP1 and mtHSP70, were significantly enriched in the insoluble fraction of the human islets of donors with T2D using immunobloting, while no accumulation of the mitochondrial chaperone HSP60 was observed (Fig. 1f,g and Extended Data Fig. 1f). We also observed that β cells of donors with T2D had more intense and punctate mtHSP70-stained areas than donors without T2D using immunofluorescence imaging, which is supportive of the presence of mitochondrial protein aggregates within β cells in T2D (Fig. 1h).
To determine if the changes in protein solubility in T2D were similar to the signatures of protein misfolding in the ER or mitochondria, we next exposed human islets from donors without T2D to the synthetic triterpenoid 2-cyano-3,12-dioxo-oleana-1,9-dien-28-oic acid methyl ester (CDDO)43,44, a well-known LONP1 inhibitor that binds its ATP-binding domain and blocks ATP hydrolysis45 to impair mitochondrial protein folding, or tunicamycin (TUN), a potent inhibitor of N-linked glycosylation of proteins in the ER to elicit ER protein misfolding. To confirm the efficacy of CDDO45,46, we observed that CDDO impaired LONP1-mediated turnover of recombinant TFAM in vitro (Extended Data Fig. 3a). TMT–MS showed that both CDDO and TUN induced profound changes in protein solubility in human islets, with CDDO eliciting a robust change in mitochondrial protein solubility (Fig. 1i–l). Enriched insoluble mitochondrial proteins after CDDO exposure included mitochondrial matrix proteins (CLPP and IDH2), mitochondrial membrane proteins (CYB5B) and OXPHOS subunits (NDUFB6 and NDUFA9), while reduced soluble proteins included ETC–OXPHOS system proteins and mitochondrial ribosome subunits (Fig. 1i,j and Extended Data Fig. 3b). As expected, TUN-exposed human islets developed an increase in insoluble proteins related to ER protein misfolding or the ER unfolded protein response, such as peptidyl-prolyl cis-trans isomerase C and the ER chaperone HSPA5 (also known as BiP), with decreases in ER luminal proteins, the ER-associated degradation component membrane (TMEM259) and glycosylated proteins, including TAP-associated glycoprotein and matrix metallopeptidase 14 (Fig. 1k,l and Extended Data Fig. 3c). Despite the well-known communication between the ER and mitochondria, we observed few changes in mitochondrial protein insolubility after TUN exposure in human islets (Fig. 1k,l). In contrast, we observed increases in ACO2, NDUFA10 and ATP5A, as well as LONP1 and mtHSP70, in the insoluble fraction of CDDO-exposed human islets without accumulation of these mitochondrial proteins in the insoluble fraction of TUN-exposed islets (Extended Data Fig. 3d,e), suggesting a similar response of ETC–OXPHOS and mitochondrial protease and chaperone solubility in human islets in T2D or after CDDO exposure. We also confirmed that CDDO elicited the accumulation of misfolded mitochondrial protein aggregates43,44,47 by observing increased immunostaining for protein aggregates and aggresomes in β cells colocalized with the mitochondrial chaperone mtHSP70, which binds to mitochondrial protein aggregates, as well as observing increases in mRNA expression of markers of the mitochondrial unfolded protein response (UPRmt) in islets, which is consistent with a transcriptional response to misfolded mitochondrial proteins (Extended Data Fig. 3f,g).
Extended Data Fig. 3. Alterations in mitochondrial protein solubility are observed in human islets following pharmacologic induction of mitochondrial rather than ER protein misfolding.
(a) TFAM and LONP1 protein levels visualized by WB (Left) and densitometry (Right) of recombinant purified human TFAM and LONP1 to assess LONP1 protease activity in the presence of 5 μM CDDO or Vehicle (DMSO). LONP1 protein levels serve as a reference/loading control. n = 3 independent experiments/group. (b) Cellular component analysis of significantly upregulated insoluble proteins (left) and cellular component analysis of significantly downregulated soluble proteins (right) from 1 μM CDDO-exposed islets compared to DMSO control islets. n = 4 independent islet donors/group. (c) Cellular component analysis of significantly upregulated insoluble proteins (left) and cellular component analysis of significantly downregulated soluble proteins (right) from 1 μg/mL tunicamycin (TUN)-exposed islets compared to DMSO control islets. n = 4 independent islet donors/group. (d) Representative WB images of selected mitochondrial proteins of human islets and (e) quantification of protein expression by densitometry (normalized to VINCULIN). n = 4 independent islet donors/group. VDAC1 serves as a soluble mitochondrial protein loading control. VINCULIN serves as a loading control for both soluble and insoluble fractions. S, soluble fraction; P, insoluble fraction. (f) Representative immunofluorescence image (n = 4/group) depicting Proteostat visualization of protein aggregates and co-localization with mtHSP70 in islets of 8-week-old C57BL/6N mice exposed to 1 μM CDDO or DMSO control for 20 h. Scale bars (Insulin, mtHSP70, Proteostat, Merge), 12.5 μm; Scale bar (zoom), 8.5 μm. Pink dashed boxes within merged image denote regions visualized at higher magnification (Zoom - far right). (g) Quantitative RT-PCR of markers of the mitochondrial unfolded protein response (UPRmt) from RNA isolated from islets of 12-week-old C57BL/6N mice exposed to 1 μM CDDO or DMSO control for 24 h. n = 3 mice/group. All data in figure are presented as mean ± SEM. Statistical analysis: 3A and 3E, *P < 0.05 by one-way ANOVA followed by Tukey’s multiple comparisons test; 3B and 3C *P < 0.05 by hypergeometric test followed by multiple hypothesis testing using FDR-corrected P values (FDR < 0.05); 3G, *P < 0.05, **P < 0.01 by unpaired two-tailed Student’s t-test.
To further visualize the intersections in insoluble protein enrichment in human islets from donors with T2D compared to pharmacological induction of mitochondrial or ER protein misfolding, we generated an UpSet plot for the T2D, CDDO and TUN groups. Interestingly, human islets from donors with T2D had a greater than fourfold increase in intersections of differently enriched insoluble proteins, with islets following CDDO exposure as opposed to TUN exposure (Fig. 1m). Together, these data support the notion that the robust change in mitochondrial protein solubility observed in human islets from donors with T2D resembles, at least in part, a signature of mitochondrial protein misfolding.
LONP1 expression is lower in β cells in T2D while β cell LONP1 deficiency results in hyperglycaemia and increased β cell apoptosis
Given our findings of a signature of mitochondrial protein misfolding in the islets of donors with T2D, we set out to profile the expression of mitochondrial matrix chaperones and proteases given their vital importance to mitochondrial proteostasis. We expected to observe an induction of LONP1 and mtHSP70 mRNA in β cells in T2D because previous work showed that these genes are induced after lipotoxicity in human and rodent islets24,31. Using single-cell RNA sequencing (scRNA-seq) data from human islet donors with or without T2D48, we were surprised to observe no significant increases in expression of mitochondrial matrix protease or chaperone genes in the β cells of islets from donors with T2D (Extended Data Fig. 4a,b). Instead, we observed a modest, yet significant downregulation of LONP1in the β cells of islets from donors with T2D (Fig. 2a). While the differences in LONP1 expression displayed donor-to-donor variability not uncommon among human islet preparations (Fig. 2a), the reductions in LONP1 expression appeared to be β-cell-specific, as we did not find lower LONP1 expression in α cells in cases of T2D (Extended Data Fig. 4a). Furthermore, we found that LONP1 protein levels were lower in the islets of donors with T2D (Fig. 2b). Of note, whereas a larger sample set of 13 human islet donors displayed a significant 35% reduction of total LONP1 protein levels in donors with T2D compared to donors without T2D (Fig. 2b) again with donor-to-donor variability, the smaller subset of islet donors with T2D profiled for the TMT–MS studies (n = 4 donors per group) displayed a 17% reduction in total LONP1 protein levels that did not reach statistical significance (Extended Data Fig. 1g).
Extended Data Fig. 4. Expression of mitochondrial proteases and chaperones is not altered in β cells and α cells of human islet donors with T2D.
Pseudobulk gene expression data, presented as log2CPM, of mitochondrial matrix proteases (a) and chaperones (b) from β cells and α-cells of human islet donors with or without T2D by single cell RNA sequencing. Box plots are presented the minimum, first quartile, median, third quartile, maximum, and interquartile range. *P < 0.05 by both unpaired two-tailed Student’s t-test and FDR < 5% for multiple testing correction. n = 17 non-diabetic donors, n = 17 donors with T2D.
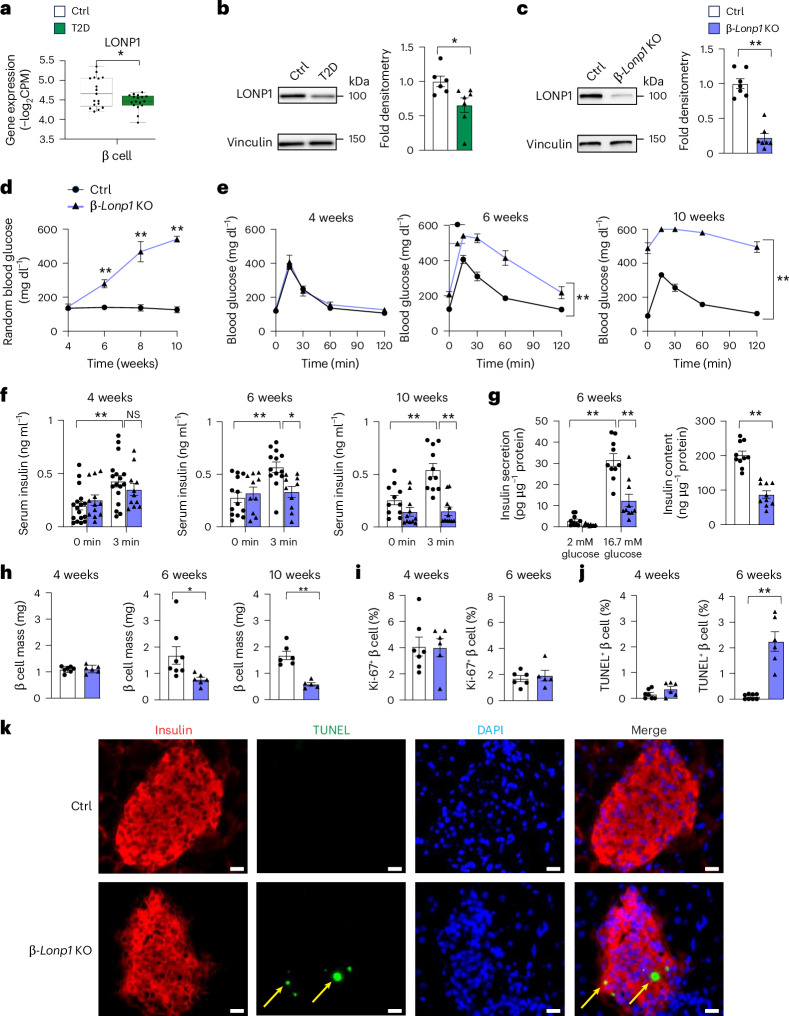
Fig. 2. Pancreatic β-cell-specific LONP1 deficiency leads to hyperglycaemia due to β cell apoptosis and loss of β cell mass.
a, Pseudobulk gene expression data of LONP1 from the β cells of human islet donors with or without T2D using scRNA-seq. n = 17 donors without T2D, n = 17 donors with T2D. The box plots present the minimum, first quartile, median, third quartile, maximum and interquartile range. b, Expression of LONP1 using immunoblotting (left) and protein densitometry (right) in human islets from donors with T2D and Ctrl donors without T2D. n = 6 donors without T2D, n = 7 donors with T2D. c, Expression of LONP1 using immunoblotting (left) and protein densitometry (right) in islets isolated from 4–6-week-old Ctrl and β-Lonp1KO mice. n = 7 mice per group. d, Random blood glucose concentrations from Ctrl and β-Lonp1 KO mice measured between the ages of 4 and 10 weeks. n = 16 Ctrl versus 12 β-Lonp1 KO at 4 weeks; n = 15 Ctrl versus 12 β-Lonp1 KO at 6 weeks; n = 6 Ctrl versus 5 β-Lonp1 KO at 8 weeks; n = 6 Ctrl versus 5 β-Lonp1 KO at 10 weeks. e, Blood glucose concentrations measured during IPGTT from Ctrl and β-Lonp1 littermates at ages 4, 6 and 10 weeks. n = 10 Ctrl versus 8 β-Lonp1 KO at 4 weeks; n = 14 Ctrl versus 11 β-Lonp1 KO at 6 weeks; n = 6 Ctrl versus 5 β-Lonp1 KO at 10 weeks. f, Serum insulin concentrations measured after in vivo glucose stimulation from Ctrl and β-Lonp1 KO mice at ages 4, 6 and 10 weeks. n = 17 Ctrl versus 11 β-Lonp1 at 4 weeks; n = 13 Ctrl versus 10 β-Lonp1 KO at 6 weeks; n = 11 Ctrl versus 11 β-Lonp1 KO at 10 weeks. g, GSIS after static incubation in 2 mM and 16.7 mM glucose (left) and islet insulin content (right), performed in isolated islets of 6-week-old Ctrl and β-Lonp1 KO littermates. n = 10 per group. h, Pancreatic β cell mass measured in Ctrl and β-Lonp1 littermates at ages 4, 6 and 10 weeks. n = 7 Ctrl versus 6 β-Lonp1 KO at 4 weeks; n = 8 Ctrl versus 6 β-Lonp1 KO at 6 weeks; n = 6 Ctrl versus 5 β-Lonp1 KO at 10 weeks. i, Quantification of β cell replication by Ki-67 and insulin immunostaining from pancreatic sections of 4-week-old and 6-week-old Ctrl and β-Lonp1 KO littermates. n = 7 Ctrl versus 6 β-Lonp1 KO at 4 weeks; n = 6 Ctrl versus 5 β-Lonp1 at 6 weeks. j, Quantification of β cell death using TUNEL and insulin immunostaining from pancreatic sections of 4-week-old and 6-week-old Ctrl and β-Lonp1 KO littermates. n = 7 Ctrl versus 6 β-Lonp1 KO at 4 weeks; n = 8 Ctrl versus 6 β-Lonp1 KO at 6 weeks. k, Representative immunofluorescence image from pancreatic sections of 6-week-old Ctrl and β-Lonp1 KO littermates for TUNEL staining. The yellow arrows indicate insulin+TUNEL+ cells. All data are presented as the mean ± s.e.m. a, *P < 0.05 was determined using both an unpaired, two-tailed Student’s t-test and FDR < 5% for multiple testing correction. b,c,h–j, *P < 0.05 and **P < 0.01 were determined using an unpaired, two-tailed Student’s t-test. d–g, *P < 0.05 and **P < 0.01 were determined using a one-way analysis of variance (ANOVA) followed by a Tukey’s multiple comparisons test. k, Scale bar, 50 μm. DAPI, 4′,6-diamidino-2-phenylindole; NS, not significant.
Our observations of reduced β cell LONP1 expression in T2D, together with increases in the insolubility and aggregation of matrix proteins in the human islets of donors with T2D similar to that of CDDO exposure, led us to query the importance of LONP1 in β cell health and function in vivo. Thus, we generated mice bearing β-cell-specific deletion of Lonp1 (Lonp1loxP/loxP;Ins1Cre, hereafter known as β-Lonp1 KO; Extended Data Fig. 5a). Lonp1loxP/loxP alone and Ins1Cre alone mice displayed no differences in glucose tolerance and body weight, which is also consistent with our observations in Ins1Cre alone mice from previous studies10,49 and were thus combined as controls (Ctrls) (Extended Data Fig. 5b,c). β-Lonp1knockout (KO) mice exhibited an ~80% reduction of LONP1 protein expression in islets when compared to littermate Ctrls (Fig. 2c).
Extended Data Fig. 5. LONP1 deficiency leads to reduced β cell mass due to increases in β cell apoptosis rather than alterations in β cell maturity/dedifferentiation.
(a) Model of Cre-mediated recombination of the LonP1 locus. (b) Ad libitum-fed body weight measured in littermate mice between ages 5–7 weeks. n = 5 Ins1+/+; LonP1loxP/loxP vs 8 Ins1Cre/+; LonP1+/+ at 5 weeks; n = 7 Ins1+/+; LonP1loxP/loxP vs 5 Ins1Cre/+; LonP1+/+ at 7 weeks. (c) Blood glucose concentrations measured during IPGTT in littermate mice at age 5 weeks (left) and age 7 weeks (right). n = 5 Ins1+/+; LonP1loxP/loxP vs 8 Ins1Cre/+; LonP1+/+ at 5 weeks; n = 7 Ins1+/+; LonP1loxP/loxP vs 5 Ins1Cre/+; LonP1+/+ at 7 weeks. (d) Blood glucose concentrations measured during IPGTT related to Fig. 2e. n = 8 male vs 6 female/Ctrl group and 7 male vs 4 female/ β-LonP1KO group. (e) Ad libitum-fed body weight measured in mice between ages 4–10 weeks. n = 10 Ctrl vs 8 β-LonP1KO at 4 weeks; n = 10 Ctrl vs 8 β-LonP1KO at 6 weeks; n = 6 Ctrl vs 5 β-LonP1KO at 10 weeks. (f) Blood glucose concentrations (presented as % of baseline glucose) measured during insulin tolerance testing (ITT) of 4-week-old (left) and 6-week-old (right) mice. n = 5 Ctrl vs 3 β-LonP1KO at 4 weeks; n = 10 Ctrl vs 6 β-LonP1KO at 6 weeks. (g) Glucose-stimulated insulin secretion following static incubations in 2 mM and 16.7 mM glucose, performed in isolated islets of 6-week-old mice (normalized to total insulin content). n = 10 mice/group. (h) Pancreatic β cell mass related to Fig. 2h. n = 3 male vs 4 female/Ctrl group and 4 male vs 2 female/ β-LonP1KO group at 4 weeks; n = 5 male vs 3 female/Ctrl group and 3 male vs 3 female/ β-LonP1KO group at 6 weeks; n = 3 male vs 3 female/Ctrl group and 4 male vs 1 female/ β-LonP1KO group at 10 weeks. (i) Quantification of cell death ELISA (normalized to total DNA content) measured in isolated islets of 6-week-old mice. n = 7 mice/group. (j) Representative immunofluorescence images (n = 5/group) depicting β cell maturity (left) or dedifferentiation markers (right) from pancreatic sections of 6-week-old mice. A representative image of pancreatic sections of high fat diet-fed of β-Clec16aKO mice as a positive control for ALDH1A3 immunostaining. Scale bars, 50 μm. All data in figure are presented as mean ± SEM. Statistical analysis: 5 G, **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons test. 5I, *P < 0.05 by unpaired two-tailed Student’s t-test.
To examine the physiological consequences of LONP1 deficiency in β cells, we first measured random blood glucose concentrations in β-Lonp1 KO mice and littermate Ctrls, observing that while β-Lonp1 KO mice were normoglycaemic shortly after weaning, they developed progressively worse hyperglycaemia beginning at 6 weeks of age (Fig. 2d). Similarly, β-Lonp1 KO mice exhibited normal glucose tolerance during an intraperitoneal glucose tolerance test (IPGTT) at 4 weeks of age, with significant and progressively worse glucose intolerance at 6 weeks and 10 weeks of age (Fig. 2e). Both male and female β-Lonp1 KO mice exhibited a similar degree of glucose intolerance at 6 weeks and 10 weeks of age compared to littermate male and female Ctrls, respectively, without overt evidence of sexually dimorphic phenotypes (Extended Data Fig. 5d). We did not observe differences in body weight or peripheral insulin sensitivity between the groups (Extended Data Fig. 5e,f). Furthermore, circulating insulin concentrations after an intraperitoneal glucose challenge in vivo progressively decreased in β-Lonp1 KO mice with age (Fig. 2f). We next assessed glucose-stimulated insulin secretion (GSIS) and insulin content in isolated islets of 6-week-old β-Lonp1 KO mice, again observing a significant reduction compared to littermate Ctrls (Fig. 2g). We next evaluated GSIS as a fraction of total insulin content and did not identify differences between the groups (Extended Data Fig. 5g), suggesting that reductions in β cell insulin release could be a consequence of reduced insulin content.
The progressive reductions in glucose tolerance and insulin release with age, coupled to reduced islet insulin content, led us to question whether the hyperglycaemia observed in β-Lonp1 KO mice was due to a decline in pancreatic β cell mass. In line with our physiological observations, β cell mass was unchanged at 4 weeks of age but it was significantly diminished in β-Lonp1 KO mice by 6 and 10 weeks of age (Fig. 2h). Both male and female β-Lonp1 KO mice exhibited a similar decline of β cell mass with age compared to littermate male and female Ctrls, respectively, again without overt evidence of sexually dimorphic phenotypes (Extended Data Fig. 5h). Pancreatic β cell mass is maintained by a balance of β cell proliferation, apoptosis and dedifferentiation or loss of identity50. To explore the aetiology of reduced β cell mass, we assessed markers of β cell proliferation and apoptosis. We first assessed β cell proliferation, observing no significant differences in β-Lonp1 KO mice at both 4 and 6 weeks of age (Fig. 2i). We next examined β cell survival using terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) in pancreatic sections of Ctrl and β-Lonp1 KO mice, noting a robust increase in the percentage of TUNEL+ β cells in 6-week-old β-Lonp1 KO mice yet no significant differences between β-Lonp1 mice and littermate Ctrls at 4 weeks of age (Fig. 2j,k). As a complementary approach to assess cell survival, we evaluated apoptosis after detection of cytoplasmic histone-bound DNA fragments using enzyme-linked immunosorbent assay (ELISA) in isolated islets of 6-week-old β-Lonp1 KO mice and Ctrl littermates, which again revealed an increase in apoptosis in LONP1-deficient β cells (Extended Data Fig. 5i). In addition, we did not observe reductions in the β cell maturity marker urocortin 3 or increases in ALDH1A3, a marker of β cell dedifferentiation51, in LONP1-deficient β cells (Extended Data Fig. 5j). Taken together, LONP1 deficiency leads to progressive hyperglycaemia and β cell failure related to impaired β cell survival and loss of β cell mass.
To ensure that the physiological and histological changes we observed were not attributable to developmental defects, we generated inducible β-cell-specific LONP1 KO animals by intercrossing our mice bearing the Lonp1 conditional allele with the tamoxifen (TMX)-inducible MIP1-CreERT strain (LonP1loxP/loxP; MIP1-CreERT, hereafter known as iβ-Lonp1 KO mice). After TMX-mediated recombination beginning at 8 weeks of age, 15-week-old iβ-Lonp1 KO mice exhibited glucose intolerance and reduced glucose-stimulated insulin release in vivo when compared to both TMX-treated MIP1-CreERT and vehicle-treated Lonp1loxP/loxP; MIP1-CreERT controls (Extended Data Fig. 6a–d). Furthermore, we observed that 15-week-old iβ-Lonp1 KO mice developed reductions in β cell mass and survival without changes in β cell replication, phenocopying constitutive β cell Lonp1 KOs (Extended Data Fig. 6e–g). These data indicate that LONP1 is vital for the preservation of postnatal β cell mass and survival that is unrelated to regulation of β cell development.
Extended Data Fig. 6. Reductions in β cell mass and glucose tolerance in adult mice following LonP1 deficiency are not due to developmental defects.
(a) Representative WB images and (b) quantification of LONP1 expression with densitometry (normalized to VINCULIN) in islets isolated from 15-week-old mice 7 weeks after Veh or TM administration. n = 4 mice/group. (c) Blood glucose concentrations measured during IPGTT in 15-week-old MIP-CreERT; LonP1loxP/loxP + TM mice (n = 15) as well as both MIP1-CreERT + TM (n = 11) and MIP-CreERT; LonP1loxP/loxP + Veh (n = 13) littermate controls 7 weeks after Veh or TM administration. (d) Serum insulin measured during in vivo glucose-stimulated insulin release testing in 15-week-old MIP-CreERT; LonP1loxP/loxP + TM mice (n = 14) as well as both MIP1-CreERT + TM (n = 11) and MIP-CreERT; LonP1loxP/loxP + Veh (n = 13) littermate controls 7 weeks after Veh or TM administration. (e) β cell mass measured in 15-week-old MIP-CreERT; LonP1loxP/loxP + TM (n = 11) as well as both MIP1-CreERT + TM (n = 8) and MIP-CreERT; LonP1loxP/loxP + Veh (n = 8) littermate controls 7 weeks after Veh or TM administration. n = 8–11 mice/group; (f) Quantification of β cell apoptosis measured as the % of TUNEL+/Insulin+ cells performed in pancreatic sections of 15-week-old MIP-CreERT; LonP1loxP/loxP + TM mice (n = 11) as well as both MIP1-CreERT + TM (n = 8) and MIP-CreERT; LonP1loxP/loxP + Veh (n = 8) littermate controls 7 weeks after Veh or TM administration. (g) Quantification of β cell replication measured as the % of Ki67+/Insulin+ cells performed in pancreatic sections of 15-week-old MIP-CreERT; LonP1loxP/loxP + TM mice as well as both MIP1-CreERT + TM and MIP-CreERT; LonP1loxP/loxP + Veh littermate controls 7 weeks after Veh or TM administration. n = 4 mice/group. All data in figure are presented as mean ± SEM. Statistical analysis: 6B–6F, *P < 0.05, **P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons test.
β cell LONP1 deficiency results in mitochondrial protein misfolding
We next examined the specific consequences of LONP1 deficiency on β cell mitochondrial function, Ca2+, membrane potential, structure, mass and protein folding. We first measured mitochondrial respiration and bioenergetics in isolated islets from 6-week-old β-Lonp1 KO mice, observing a potent reduction in glucose-stimulated oxygen consumption and ATP concentrations compared to littermate Ctrl islets (Fig. 3a and Extended Data Fig. 7a). Given the importance of mitochondrial function for cytosolic and mitochondrial Ca2+ homeostasis in β cells52, we simultaneously measured cytosolic and mitochondrial [Ca2+] at subcellular resolution after loading with the cytosolic Ca2+ indicator Cal-520 and after adenoviral transduction with the genetically encoded mitochondrial Ca2+ indicator, mito-R-GECO, in isolated islets of β-Lonp1 KO mice and littermate Ctrls (Extended Data Fig. 7b). After glucose stimulation, real-time confocal imaging revealed the expected increase in both cytosolic and mitochondrial [Ca2+] in Ctrl islets and individual β cells (Extended Data Figs. 7c,f and 8a). However, compared to Ctrl islets, LONP1-deficient islets and β cells displayed reductions in glucose-stimulated cytosolic [Ca2+], while mitochondrial [Ca2+] changes remained unaltered (Extended Data Figs. 7c,f and 8b). Furthermore, after exposure to the uncoupler carbonyl cyanide-4-phenylhydrazon (FCCP), we observed a significant lowering in mitochondrial membrane potential (Δψm) in control islets, whereas we did not observe further lowering in Δψm upon FCCP exposure in β-Lonp1 KO islets. The latter finding is suggestive of intrinsic impairments in Δψm in the mitochondria of LONP1-deficient mice that could not be further depolarized (Extended Data Fig. 8c,d). Together, these results support the importance of LONP1 in the maintenance of β cell mitochondrial function, bioenergetics and membrane potential while not altering mitochondrial [Ca2+].
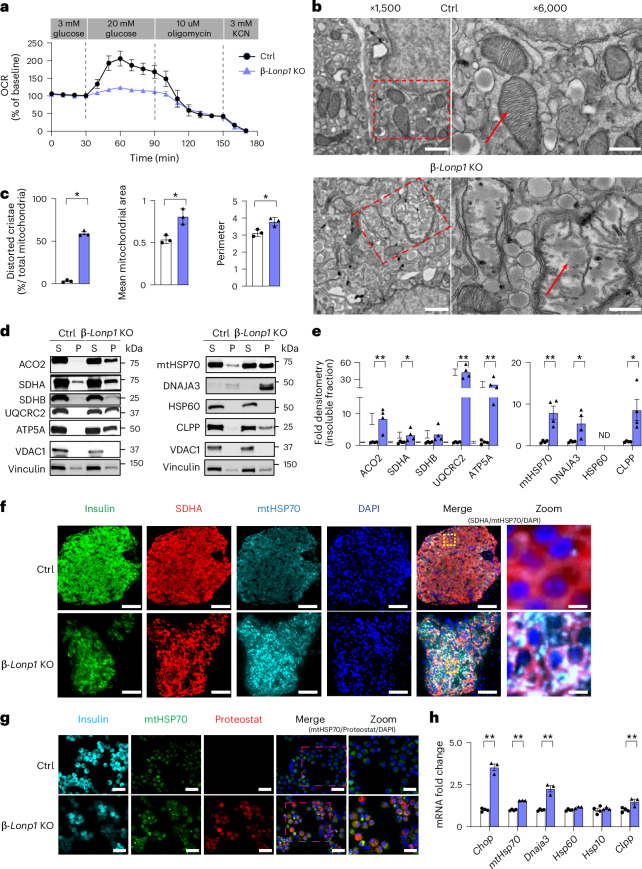
Fig. 3. Deficiency of LONP1 results in impaired mitochondrial respiration, accumulation of misfolded mitochondrial proteins and activation of the UPRmt in β cells.
a, Oxygen consumption rate (OCR) measured after exposure to 3 mM glucose, 20 mM glucose, 10 μM oligomycin and 3 mM KCN in isolated islets from 6-week-old littermate Ctrl and β-Lonp1 KO mice. n = 3 mice per group. b, Representative TEM images from the β cells of 6-week-old Ctrl and β-Lonp1 KO mice. The red rectangle with the dashed outline on the left highlights the focused area of mitochondria on the right. The red arrow denotes mitochondria. n = 3 mice per group. c, Quantification of TEM images of mitochondria (~100 independent mitochondria scored per animal) with distorted cristae, mitochondrial area and mitochondrial perimeter in the β cells of 6-week-old Ctrl and β-Lonp1 KO mice. n = 3 per group. d,e, Representative immunoblotting images (d) and quantification of mitochondrial insoluble proteins (normalized to vinculin) (e) from 6-week-old Ctrl and β-Lonp1 KO mice. VDAC1 serves as a soluble mitochondrial protein loading control. Vinculin serves as a loading control for both soluble and insoluble fractions. n = 4 per group. f, Representative immunofluorescence image (n = 4 per group) depicting mtHSP70 expression and localization from pancreatic sections of 6-week-old Ctrl mice and β-Lonp1 littermates. The yellow boxes in the merged image are visualized at higher magnification (zoom, far right). g, Representative immunofluorescence image (n = 3 per group) depicting Proteostat visualization of protein aggregates and colocalization with mtHSP70 in the islets of 6-week-old Ctrl mice and β-Lonp1 KO littermates. The pink dashed boxes in the merged image denote regions visualized at higher magnification (zoom, far right). h, Quantitative PCR with reverse transcription (RT–qPCR) of markers of the UPRmt from RNA isolated from 6-week-old Ctrl and β-Lonp1 KO islets. n = 4 Ctrl versus 3 β-Lonp1 KO. All data are presented as the mean ± s.e.m. c,e,h, *P < 0.05 and **P < 0.01 were determined using an unpaired, two-tailed Student’s t-test. b, Scale bar (left), 2 μm; scale bar (right), 500 nm. f, Scale bar (insulin, SDHA, mtHSP70, DAPI, merge), 50 μm; scale bar (zoomed image), 6.25 μm. g, Scale bar (insulin, mtHSP70, Proteostat, merge), 12.5 μm; scale bar (zoomed image), 8.5 μm.
Extended Data Fig. 7. LONP1 deficient β-cells display diminished glucose-stimulated bioenergetics and cytosolic Ca2+ without changes in mitochondrial Ca2+.
(a) Relative ATP levels (normalized to total protein) in islets isolated from 6-week-old mice following exposure to 2 mM glucose (2G) and 17 mM glucose (17G) stimulation. n = 5 Ctrl vs 3 β-LonP1KO mice; *P < 0.01 by one-way ANOVA followed by Tukey’s multiple comparisons test. (b) Representative fluorescence images of Ca2+uptake in islets from 8-week-old control (Ctrl) (i–iv) and β-LonP1KO (v–viii) littermates transduced with the mitochondrial Ca2+ indicator mito-R-Geco (purple) and loaded with the cytosolic Ca2+ indicator Cal520 (green) at indicated time points. Scale bar, 10 μm. Subsequent analyses were performed by capturing images across the whole islet (c, d), or only in those individual cells where both Cal520 and R-Geco fluorescence were detectable initially (e, f). (c) Whole islet [Ca2+]cyt changes and corresponding area under curve (AUC) in response to 3, 11 and 17 mmol/l glucose (3G, 11G and 17G) and 20 mmol/l KCl following Cal520 uptake. Traces represent mean normalized fluorescence intensity over time (F/Fmin). n = 12 islets/group (4 mice/group). 11 G AUC measured between time 3–18 min, 17 G AUC measured between time 18–33 min. (d) Whole islet [Ca2+]mito changes and corresponding AUC in response to 3, 11 and 17 mmol/l glucose and 20 mmol/l KCl following mito-R-Geco transduction. n = 12 islets/group (4 mice/group). (e) [Ca2+]cyt and (f) [Ca2+]mito dynamics and corresponding AUC from individual cells for each islet in response to 3, 11 and 17 mmol/l glucose and 20 mmol/l KCl. Traces represent mean normalized fluorescence intensity over time (F/Fmin). n = 84 cells/Ctrl group, n = 41 cells/ β-LonP1KO group (4 mice/group). 11 G AUC measured between time 3–18 min, 17 G AUC measured between time 18–33 min. All data in figure are presented as mean ± SEM. Statistical analysis: 7 C and 7E, *P < 0.05, **P < 0.01 by unpaired two-tailed Student’s t-test.
Extended Data Fig. 8. Examination of relative single cell cytosolic and mitochondrial Ca2+ concentrations as well as mitochondrial membrane potential following β-cell LONP1 deficiency.
(a) [Ca2+]cyt (left) and [Ca2+]mito (right) changes were simultaneously assessed in individual cells from the same control islet following cytosolic Cal520 uptake and mito-R-Geco transduction in response to 3, 11, and 17 mM glucose (3 G, 11 G and 17 G) and 20 mM KCl. (n = 5 cells, each color represents one cell). (b) [Ca2+]cyt (left) and [Ca2+]mito (right) changes were simultaneously assessed in individual cells from the same β-LonP1KO islet following cytosolic Cal520 uptake and mito-R-Geco transduction in response to 3, 11, and 17 mM glucose (3 G, 11 G and 17 G) and 20 mM KCl (n = 4 cells, each color represents one cell). (c) Δψm measured in dissociated β-cells loaded with 10 nM TMRM following exposure to 3- and 17-mM glucose (3 G and 17 G), or 1 µM FCCP. Traces represent normalized fluorescence intensity over time (F/Fmin) n = 120 islets/group (3 mice/group). Pink dashed box highlights changes in Δψm before and after FCCP exposure. (d) Change in Δψm post FCCP, measured as the difference between Δψm from time 13–13.5 min (after 1 µM FCCP exposure) and time 12.5–13 min (prior to FCCP) corresponding to pink dashed box in Extended Data Fig. 8c. Each dot represents an individual cell. n = 103 cells/Ctrl group, n = 104 cells/ β-LonP1KO group (3 mice/group). All data in figure are presented as mean ± SEM. Statistical analysis: 10D, **P < 0.01 by unpaired two-tailed Student’s t-test.
We next used transmission electron microscopy (TEM) and high-resolution three-dimensional (3D) deconvolution immunofluorescence imaging to examine mitochondrial ultrastructure and morphology, and networking, respectively. We observed using TEM that mitochondria in β cells from β-Lonp1 KO mice developed severely distorted cristae and increased area, as well as alterations in matrix density, which could be suggestive of an accumulation of protein aggregates (Fig. 3b,c). Consistent with the observations made using TEM, measures of mitochondrial morphology and networking analysed using high-resolution 3D deconvolution imaging also revealed greater mitochondrial volume and surface area and lower sphericity in LONP1-deficient β cells compared to littermate Ctrls, without overt defects in networking (Extended Data Fig. 9a,b).
Extended Data Fig. 9. LonP1 deficiency leads to abnormal mitochondrial morphology and a decline of mitochondrial mass.
(a) Imaris® generated three-dimensional reconstruction of deconvolution immunofluorescence Z-stack images at 60x magnification in pancreatic sections of 6-week-old mice. Each unique color represents a separate β cell mitochondrial network cluster. (b) β cell mitochondrial morphology and network analysis of deconvolution immunofluorescence Z-stack images at 60X magnification from pancreatic sections of 6-week-old mice by Mitochondria Analyzer. n = 6 mice/group. (c) Quantitative RT-PCR of markers of ER stress from RNA isolated from 6-week-old islets. n = 4 Ctrl vs 3 β-LonP1KO mice. (d) Representative WB images and (e) quantification of OXPHOS complex subunits and TOM20 in isolated islets from mice at both 4-weeks and 6-weeks of age. n = 7 Ctrl vs 7 β-LonP1KO mice at 4 weeks; n = 5 Ctrl vs 5 β-LonP1KO mice at 6 weeks. (f) Relative mtDNA content normalized to nuclear DNA expression measured by qPCR in isolated islets of mice at both 4-weeks (left) and 6-weeks (right) of age. n = 4 Ctrl vs 4 β-LonP1KO mice at 4 weeks; n = 5 Ctrl vs 4 β-LonP1KO mice at 6 weeks. (g) Citrate synthase activity measured in isolated islets of mice at both 4-weeks (left) and 6-weeks (right) of age. n = 3 Ctrl vs 3 β-LonP1KO mice at 4 weeks; n = 4 Ctrl vs 3 β-LonP1KO mice at 6 weeks. (h) Representative images (left) and quantification (right) of LONP1 levels in Min6 β-cells 72 h after transfection with siLonP1 or siCtrl. n = 6 independent experiments/group. (i) Representative images (left) and quantification (right) of phospho-γH2AX expression in isolated islets of 4-week-old mice. n = 4 mice/group. All data in figure are presented mean ± SEM. Statistical analysis: 9B, 9 C, 9E, 9 F, 9 G, and 9H, *P < 0.05, **P < 0.01 by unpaired two-tailed Student’s t-test.
As LONP1 is vital for protein quality control, and TEM of mitochondria from β cells of LONP1-deficient mice showed accumulation of protein aggregates, we next examined mitochondrial protein solubility. Indeed, we observed greater levels of several insoluble proteins in the ETC–OXPHOS system in islets isolated from 6-week-old β-Lonp1 KO mice (Fig. 3d,e). We also observed an accumulation of mtHSP70 and its co-chaperone DNAJA3 in the insoluble fraction of 6-week-old β-Lonp1 KO islets, and the mitochondrial protease CLPP (Fig. 3d,e). Furthermore, immunofluorescence imaging showed that LONP1-deficient β cells had more intense and punctate mtHSP70-stained areas than Ctrls, suggestive of the presence of mitochondrial protein aggregates (Fig. 3f). We also visualized increased immunostaining for mitochondrial protein aggregates in β cells colocalized with mtHSP70 in LONP1-deficient β cells (Fig. 3g). Accordingly, we observed increases in mRNA expression of several markers of UPRmt (Fig. 3h), which was consistent with a transcriptional response to misfolded mitochondrial proteins in LONP1-deficient β cells. In contrast, only a modest upregulation of Ire1α expression was found; no increases in expression of other ER stress markers were observed (Extended Data Fig. 9c). Together, these results suggest that a specific defect in mitochondrial protein folding occurs in β-Lonp1 KO islets.
Notably, LONP1 regulates the stability of the ETC–OXPHOS complex25,28. Thus, we examined the expression of subunits of all five OXPHOS complexes, as well as the outer mitochondrial membrane protein TOM20 (a common marker of mitochondrial mass), using immunoblotting in the islets of littermate Ctrl and β-Lonp1 KO mice. Interestingly, a variety of changes in OXPHOS subunits were observed in islets from 4-week-old β-Lonp1 KO mice, including decreases of the complex I subunit NDUFB8 and complex IV subunit MTCO1, while the complex III subunit UQCRC2 was increased (Extended Data Fig. 9d,e). However, at 6 weeks of age β-Lonp1 KO islets developed significant reductions of all subunits of all five OXPHOS complexes as well as TOM20, suggestive of a reduction in mitochondrial mass (Extended Data Fig. 9d,e). We next assessed whether mitochondrial mass was altered after loss of LONP1 in β cells using several other complementary approaches. We observed a slight reduction in mtDNA content in the islets from 4-week-old β-Lonp1 KO mice, which was amplified at 6 weeks of age (Extended Data Fig. 9f). Furthermore, citrate synthase activity was significantly lower in the islets of 6-week-old β-Lonp1 KO mice compared to littermate Ctrls, while no difference was observed at 4 weeks of age (Extended Data Fig. 9g). These results are suggestive of early alterations of ETC–OXPHOS stability followed by a later loss of mitochondrial mass. Taken together, our results demonstrate a key role for LONP1 in the maintenance of mitochondrial proteostasis in β cells, which preserves mitochondrial respiration, the ETC–OXPHOS machinery and mitochondrial mass.
Scavenging of ROS induced by LONP1 deficiency transiently improves β cell survival and glucose homeostasis
We found a striking impairment in β cell survival after LONP1 deficiency, yet the aetiology of β cell apoptosis was unclear. Mitochondrial dysfunction has been broadly associated with increases in the generation of free radicals that can lead to DNA damage and cell death53. Furthermore, LONP1 deficiency was recently observed to induce ROS and DNA damage as a key mediator of cardiomyocyte demise54. ROS is a known inducer of β cell apoptosis and reported to be elevated in the human islets of donors with T2D13. Historically, β cells have low antioxidant capacity, which sensitizes them to oxidative damage, although more recent work has challenged this belief55,56. Thus, we wished to determine if the generation of oxidative stress and DNA damage may be a driver of β cell apoptosis in β-Lonp1 KO mice. We observed an increase in ROS levels in 6-week-old β-Lonp1 KO islets compared to Ctrls using flow cytometry (Fig. 4a). We next used immunoblotting to evaluate the phosphorylation of γH2AX, a marker of DNA double-strand breaks that can be increased in the setting of oxidative stress, which was also elevated in the islets of β-Lonp1 KO mice (Fig. 4b).
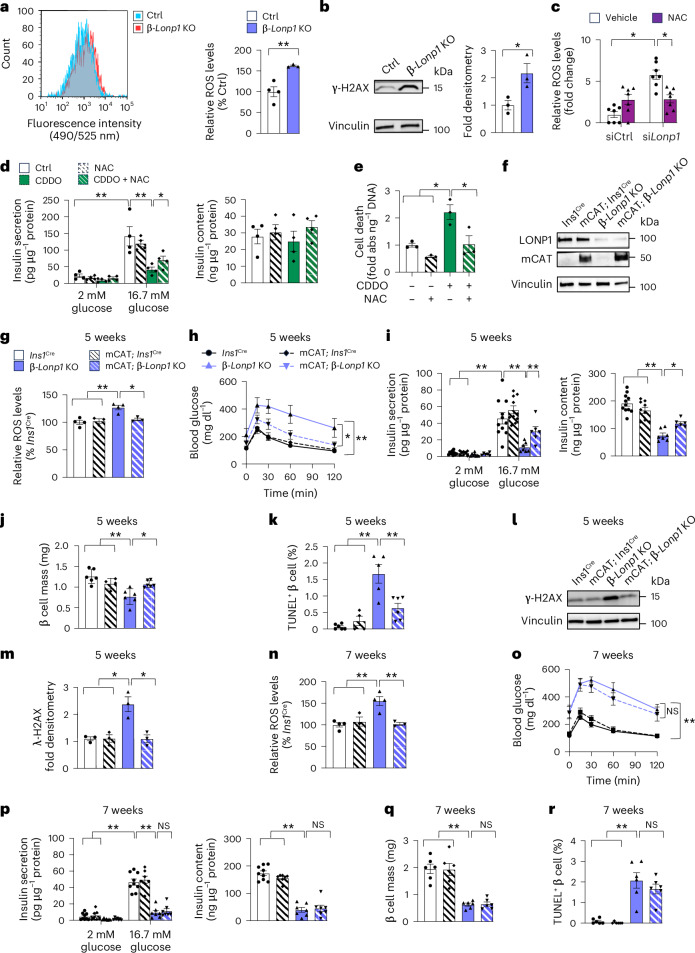
Fig. 4. Genetic or pharmacological free radical scavengers provide transiently improved cell survival after LONP1 deficiency in mouse and human islets.
a, Representative flow cytometry histogram demonstrating cellular ROS (left) and quantification of relative ROS levels (right) in dispersed islets isolated from 6-week-old mice. n = 4 Ctrl versus 3 β-Lonp1 KO. b, Representative immunoblot images (left) and densitometry (right) of phospho-γH2AX in islets isolated from 6-week-old mice. n = 3 mice per group. c, Quantification of relative ROS levels in Min6 β cells 72 h after transfection with siLonp1 or siCtrl and treated with 5 mM NAC or vehicle Ctrl for the final 36 h. n = 7 per group. d, GSIS (left) after static incubation in 2 mM and 16.7 mM glucose, and islet insulin content (right), measured in human islets treated with or without 1 µM CDDO and with or without 5 µM NAC for 24 h. n = 4 independent islet donors per group. e, Quantification of cell death measured in human islets treated with or without 1 µM CDDO and with or without 5 mM NAC for 24 h. n = 4 independent islet donors per group. f, Expression of catalase and LONP1 using immunoblotting in islets isolated from 5–7-week-old mice. Representative of three mice per group. g, Quantification of relative ROS levels determined using flow cytometry in the isolated islets of 5-week-old mice. n = 3 mice per group. h, Blood glucose concentrations measured during IPGTT from 5-week-old Ins1Cre (n = 15), mCAT; Ins1Cre (n = 6), β-Lonp1 KO (n = 8) and mCAT; β-Lonp1 KO mice (n = 13). i, GSIS (left) after static incubation in 2 mM and 16.7 mM glucose, and islet insulin content (right) measured in isolated islets of 5-week-old Ins1Cre (n = 11), mCAT; Ins1Cre (n = 10), β-Lonp1 KO (n = 7) and mCAT; β-Lonp1 KO (n = 6) mice. j, Pancreatic β cell mass measured in 5-week-old Ins1Cre (n = 6), mCAT; Ins1Cre (n = 4), β-Lonp1 KO (n = 6) and mCAT; β-Lonp1 KO (n = 6) mice. k, Quantification of β cell death using TUNEL and insulin immunostaining from pancreatic sections of 5-week-old Ins1Cre (n = 6), mCAT; Ins1Cre (n = 4), β-Lonp1 KO (n = 5) and mCAT; β-Lonp1 KO (n = 6). l, Expression of phospho-γH2AX using immunoblotting. m, Phospho-γH2AX densitometry in islets isolated from 5-week-old mice. n = 3 mice per group. n, Quantification of relative ROS production in the islets of 7-week-old mice. n = 3 mice per group. o, Blood glucose levels measured during IPGTT from 7-week-old Ins1Cre (n = 11), mCAT; Ins1Cre (n = 4), β-Lonp1 KO (n = 7) and mCAT; β-Lonp1 KO (n = 9) mice. p, GSIS (left) after static incubation in 2 mM and 16.7 mM glucose, and islet insulin content (right) measured in the isolated islets of 7-week-old Ins1Cre (n = 9), mCAT; Ins1Cre (n = 7), β-Lonp1 KO (n = 7) and mCAT; β-Lonp1 KO (n = 7) mice. q, β cell mass determined in pancreatic sections of 7-week-old mice. n = 6 mice per group. r, Quantification of TUNEL staining performed in pancreatic sections of 7-week-old mice for β cell apoptosis. n = 6 mice per group. All data are presented as the mean ± s.e.m. a,b, *P < 0.05 and **P < 0.01 were determined using an unpaired, two-tailed Student’s t-test. c–e,g–k,m–r, *P < 0.05 and **P < 0.01 were determined using a one-way ANOVA followed by Tukey’s multiple comparisons test.
To determine if ROS contributes to β cell failure after LONP1 deficiency, we used both pharmacological and genetic approaches to ameliorate oxidative stress. We first confirmed that treatment with the antioxidant N-acetylcysteine (NAC) relieved the elevated ROS levels in Min6 β cells after siRNA-mediated Lonp1 knockdown (Fig. 4c and Extended Data Fig. 9h). We next exposed human islets to CDDO, observing reductions in GSIS and cell survival similar to those present in β-Lonp1 KO islets, and found that treatment with NAC ameliorated CDDO-mediated defects in GSIS and cell survival (Fig. 4d,e). To test if excess mitochondrial ROS (mtROS) led to β cell dysfunction after LONP1 deficiency in vivo, we intercrossed β-Lonp1 KO mice (or Ins1Cre Ctrls) with Cre-inducible mitochondrial-targeted human catalase (mCAT) overexpression mice to selectively scavenge β cell mtROS because mCAT overexpression reduces H2O2and superoxide in β cells57,58. Human catalase was detected using a human-specific catalase antibody in the islets of mCAT; β-Lonp1 KO mice along with continued efficient deletion of Lonp1 (Fig. 4f), which was similar to the Lonp1 deletion efficiency we observed in Fig. 2c. Overexpression of mCAT in LONP1-deficient mice restored islet ROS to levels similar to those of Ins1Cre and mCAT; Ins1Cre Ctrls (Fig. 4g). Overexpression of mCAT improved glucose intolerance, glucose-stimulated insulin release, β cell mass and β cell survival after LONP1 deficiency at 5 weeks of age (Fig. 4h–k). In addition, overexpression of mCAT resulted in lower levels of phosphorylated γH2AX after LONP1 deficiency (Fig. 4l,m). However, the beneficial effects of mCAT overexpression in LONP1-deficient mice were short-lived; mCAT overexpression did not sustain improvements in glucose tolerance, serum insulin levels, β cell mass and β cell survival by 7 weeks of age, despite continued improvements in ROS levels (Fig. 4n–r). Together, these observations suggest that while reductions in ROS elicit an acute and transient protective effect on β cell survival, oxidative stress is unlikely to be the primary mediator of β cell demise after LONP1 deficiency.
Mitochondrial protein misfolding precedes oxidative stress after LONP1 deficiency
Our observations of mitochondrial protein misfolding and oxidative stress after LONP1 deficiency next led us to determine if ROS induced mitochondrial protein misfolding. We first evaluated soluble and insoluble protein fractions in human islets exposed to CDDO, observing an accumulation of ETC–OXPHOS proteins, as well as the mtHSP70 chaperone machinery and LONP1 itself in the insoluble fraction (Fig. 5a,b). After NAC treatment, we did not observe improvements in mitochondrial protein misfolding in human islets exposed to CDDO (Fig. 5a,b). We next evaluated mitochondrial protein misfolding in islets from β-Lonp1 KO mice and LONP1-deficient mice bearing mCAT overexpression. Similar to observations in human islets, we found that β cell mCAT overexpression in mice was unable to relieve misfolding of ETC–OXPHOS proteins or the presence of the mtHSP70 machinery and CLPP in the insoluble fraction in LONP1-deficient β cells both at 5 and 7 weeks of age (Fig. 5c,d). These results suggest that β cell mitochondrial protein misfolding after LONP1 deficiency is not a consequence of oxidative stress. These results also led us to speculate that the continued presence of mitochondrial protein misfolding in mCAT; β-Lonp1 KO mice may override the transient protective effects of antioxidant exposure, ultimately leading to hyperglycaemia and loss of β cell mass.
Fig. 5. Mitochondrial protein misfolding is not ameliorated by antioxidants and precedes the appearance of oxidative stress after LONP1 deficiency.
a, Representative immunoblotting images of selected mitochondrial proteins. b, Quantification of mitochondrial insoluble proteins (normalized to vinculin) using the densitometry of human islets exposed to dimethyl sulfoxide (DMSO) (Ctrl), 1 μM CDDO or 1 μM CDDO + 5 mM NAC for 24 h. VDAC1 serves as a soluble mitochondrial protein loading control. Vinculin serves as a loading control for both soluble and insoluble fractions. n = 4 independent human islet donors per group. c, Representative immunoblot images of ETC–OXPHOS system proteins (top) and quantification of mitochondrial insoluble proteins (normalized to vinculin) using densitometry (bottom) in the isolated islets of mice at both 5 and 7 weeks of age. n = 3 biological replicates per group. d, Representative immunoblot images of mitochondrial matrix chaperones and proteases (top) and quantification of fractions of mitochondrial insoluble proteins (normalized to vinculin) using densitometry (bottom) of isolated islets of mice at both 5 and 7 weeks of age. n = 3 biological replicates per group. e, Representative immunoblot images of ETC–OXPHOS system proteins (left) and quantification of mitochondrial insoluble proteins (normalized to vinculin) using densitometry (right) of the isolated islets of 4-week-old mice. n = 3 independent mice per group. f, Representative immunoblot images of mitochondrial chaperones and proteases (left) and quantification of mitochondrial insoluble proteins (normalized to vinculin) using densitometry (right) of the isolated islets of 4-week-old mice. n = 3 independent mice per group. g, Quantification of relative ROS production in the islets of 4-week-old mice. n = 3 mice per group. h, BN-PAGE followed by immunoblotting for OXPHOS complexes performed in the isolated islets from 4-week-old mice. The quantification of complexes I, III, IV and V (normalized to complex II) using densitometry from BN-PAGE studies is shown in the graph on the right. n = 4 per group. All data are presented as the mean ± s.e.m. b–d, *P < 0.05 and **P < 0.01 were determined using a one-way ANOVA followed by Tukey’s multiple comparisons test. e,f,h, *P < 0.05 and **P < 0.01 were determined using an unpaired, two-tailed Student’s t-test.
To better clarify the chronological aetiology of mitochondrial dysfunction leading sequentially to β cell failure after deficiency of LONP1, we assessed key mitochondrial defects visible in β-Lonp1 KO mice at 4 weeks of age. As shown above, no differences in glycaemic control, glucose-stimulated insulin release, β cell mass, β cell survival or mitochondrial mass were visible in β-Lonp1 KO mice compared to littermate Ctrls at this age (Fig. 2d–f,h,j and Extended Data Fig. 9g), thus allowing us to identify mechanistic defects that are forerunners of β cell dysfunction and are not consequences of hyperglycaemia or glucotoxicity. Interestingly, we observed evidence of mitochondrial protein misfolding in the β-Lonp1 KO islets of 4-week-old mice, including increases in insoluble ETC–OXPHOS proteins, as well as the mtHSP70 chaperone machinery and CLPP (Fig. 5e,f). Notably, these increases in insoluble mitochondrial proteins developed after LONP1 deficiency despite no differences in islet ROS levels or oxidative DNA damage at this age (Fig. 5g and Extended Data Fig. 9i).
Our observations of insoluble ETC–OXPHOS subunits preceding hyperglycaemia in β-Lonp1 KO mice next led us to query if formation of fully assembled ETC complexes was impaired. Indeed, loss of key subunits of complexes I, III and IV in β cells all led to hyperglycaemia59. Thus, we examined ETC complexes using Blue Native polyacrylamide gel electrophoresis (BN-PAGE). Importantly, complexes I, III, IV and V were all lower in the islets of 4-week old β-Lonp1 KO mice (Fig. 5h) before changes in mitochondrial mass (Extended Data Fig. 9d–g). Taken together, these observations suggest that mitochondrial protein misfolding, not oxidative stress, is the initial insult detected after LONP1 deficiency, which heralds the development of ETC–OXPHOS system defects, β cell failure and hyperglycaemia.
LONP1-mtHSP70 chaperone activity promotes β cell survival
As prior work supported the concept that LONP1 acts as a protease or as an ATP-dependent chaperone, functioning independently of its protease activity29,30, we wished to clarify the specific mechanism according to which LONP1 acts to prevent mitochondrial protein misfolding and maintain β cell survival. To determine if LONP1 requires protease activity to promote β cell survival, we expressed a protease-deficient Lonp1S855A mutant25,29 or an empty vector (EV) control in the β cells of the islets of 5-week-old β-Lonp1 KO mice (or littermate Ctrls) using the pseudoislet approach49. Briefly, primary islets were dispersed, transduced with adenoviral particles encoding Lonp1S855A or EV under control of the rat insulin 2 promoter (Ad.RIP2.Lonp1S855A or Ad.RIP2.EV, respectively) to facilitate β-cell-specific expression and then reaggregated into pseudoislets (Fig. 6a). We then assessed β cell survival 7 days after pseudoislet generation after dissociation and cytocentrifugation onto slides for TUNEL analysis. Importantly, we found that re-expression of the protease dead Lonp1S855A mutant significantly rescued β cell apoptosis in β-Lonp1 KO pseudoislets (Fig. 6b,c). As an additional control, we also confirmed that overexpression of the Lonp1S855A mutant lacked the ability of LONP1 to reduce expression of its protease substrates HMGCS2 (ref. 60) and Twinkle61 in Min6 β cells (Extended Data Fig. 10a).
Fig. 6. LONP1-mtHSP70 chaperone activity promotes β cell survival by relieving mitochondrial protein misfolding.
a, Schematic diagram illustrating the generation of mouse pseudoislets. b, TUNEL staining for β cell apoptosis performed in mouse pseudoislets dissociated for cytocentrifugation from 5-week-old Ctrl mice and β-Lonp1 KO littermates performed 7 days after adenoviral transduction with RIP2-driven EV (Ad.RIP2.EV) and the protease-deficient Lonp1S855A mutant (Ad.RIP2.Lonp1S855A), with exposure to vehicle (DMSO) or 1 μM of the mtHSP70 inhibitor MKT077 for the final 24 h. Representative images of 3–4 mice per group. The yellow arrows indicate insulin+TUNEL+ cells. c, Quantification of TUNEL staining from the studies in b. n = 4 vehicle versus three MKT077 biological replicates. d, Expression of mitochondrial proteins from the soluble and insoluble fractions performed in mouse pseudoislets from 5-week-old Ctrl and β-Lonp1 KO mice 7 days after adenoviral transduction with Ad.RIP2.EV or Ad.RIP2.Lonp1S855A using immunoblotting. Representative images of three mice per group. VDAC1 serves as a soluble mitochondrial protein loading control. Vinculin serves as a loading control for both soluble and insoluble fractions. e, Quantification of mitochondrial insoluble proteins (normalized to vinculin) from the studies in d. n = 3 biological replicates per group. f, Expression of mitochondrial proteins from soluble and insoluble fractions of mouse pseudoislets generated from 8-week-old Lonp1loxP/loxP; MIP1-CreERT mice 7 days after adenoviral transduction with Ad.RIP2.EV or Ad.RIP2.Lonp1S855A. Pseudoislets were co-cultured with vehicle (EtOH) or 2 μM 4-hydroxytamoxifen (4-OHT) to induce recombination in vitro and generate experimental groups (Ctrl or iβ-Lonp1 KO, respectively) before the generation of soluble or insoluble fractions for immunoblotting. Representative images of three biological replicates per group. g, Quantification of mitochondrial insoluble proteins (normalized to vinculin) from the studies in f. n = 3 biological replicates per group. All data are presented as the mean ± s.e.m. c,e,g, *P < 0.05 and **P < 0.01 were determined using a one-way ANOVA followed by Tukey’s multiple comparisons test. b, Scale bar, 50 μm.
Extended Data Fig. 10. Examination of LONP1 activity, interaction with mtHSP70, pharmacologic activation, and transcriptional regulation in pancreatic β-cells.
(a) Representative WB images (left) and quantification (right) of FLAG-epitope tagged LONP1, total LONP1, HMGCS2, and TWINKLE expression in Min6 β-cells 72 h after transfection with pQCXIP vectors expressing a FLAG-tagged empty vector (EV), wild-type LONP1 (WT), or LONP1 S855A mutant (S855A). VINCULIN serves as a loading control. n = 3 independent experiments/group. (b) Representative WB of lysates of Min6 β-cells following control anti-IgG immunoprecipitation (IP; middle lane) or anti-LONP1 IP (right lane). n = 4 independent experiments/group. (c) Aconitase activity measured in Min6 β-cells exposed to 0.3 μM MKT077 or DMSO for 24 h. n = 3/group. (d) TFAM and LONP1 protein levels visualized by WB (Left) and densitometry (Right) of recombinant purified human TFAM and LONP1 to assess LONP1 protease activity in the presence of 40 μM 84-B10 or vehicle control (DMSO). LONP1 protein levels serve as a reference/loading control. n = 3 independent experiments/group. (e) Immunofluorescence imaging performed in human β-cell enriched pseudoislets, generated by magnetic sorting for the β cell surface marker NTPDase3, following dissociation for cytocentrifugation and imaging, stained for insulin (red) and DAPI (DNA - blue). Scale bars, 50 μm. Representative image of 4 β-cell enriched pseudoislet preparations each from independent human islet donors. (f) Pseudobulk gene expression data of reported transcriptional regulators of LONP1 from β cells of human islet donors with or without T2D by single cell RNA sequencing. Box plots are presented the minimum, first quartile, median, third quartile, maximum, and interquartile range. n = 17 non-diabetic donors, n = 17 donors with T2D. All data in figure are presented mean ± SEM. Statistical analysis: 10 A, 10D, *P < 0.05 by one-way ANOVA followed by Tukey’s multiple comparisons test. 10 C, *P < 0.05 by unpaired two-tailed Student’s t-test.
Given a previous report displaying the partnership between LONP1 and mtHSP70 necessary for LONP1 chaperone-like activity29, we next tested the importance of LONP1-mtHSP70 function in β cells. We first observed that LONP1 interacted with mtHSP70 using immunoprecipitation assays in Min6 β cells (Extended Data Fig. 10b). Next, we exposed β-Lonp1 KO pseudoislets to MKT077, an allosteric inhibitor of mtHSP70 (ref. 62), to determine if the beneficial effects of Lonp1S855A re-expression were related to LONP1-mtHSP70 function. As a Ctrl, we first confirmed that exposure to MKT077 reduced aconitase activity in Min6 β cells, which is consistent with the importance of mtHSP70 to maintain proper folding of aconitase63 (Extended Data Fig. 10c). Interestingly, MKT077 exposure abrogated the rescue of Ad.RIP2.Lonp1S855A on cell survival in β-Lonp1 KO pseudoislets, suggesting that LONP1-mtHSP70 chaperone activity is necessary to maintain β cell survival (Fig. 6b,c).
We further observed that Ad.RIP2.Lonp1S855A transduction significantly reduced insoluble ETC–OXPHOS proteins, as well as the mtHSP70 chaperone machinery and CLPP in β-Lonp1 KO pseudoislets (Fig. 6d). However, there was only a partial rescue of mitochondrial protein misfolding 7 days after Ad.RIP2.Lonp1S855A transduction in the islets of 5-week-old constitutive β-Lonp1 mice (Fig. 6d,e). This led us to speculate that the prolonged burden of misfolded mitochondrial proteins could not be completely cleared within 1 week of Lonp1S855A re-expression. Thus, to elicit acute formation of misfolded mitochondrial proteins because of loss of LONP1, we next generated pseudoislets from iβ-Lonp1 KO mice after in vitro recombination achieved using culture in the presence of 4-OHT (or vehicle control). We again observed increases in ETC–OXPHOS proteins and the mtHSP70 chaperone machinery in the insoluble fraction of iβ-Lonp1 KO pseudoislets (Fig. 6f,g). Importantly, transduction of iβ-Lonp1 KO pseudoislets with Ad.RIP2.Lonp1S855A reversed the accumulation of misfolded mitochondrial proteins (Fig. 6f,g), which is consistent with a key protease-independent role for LONP1 to maintain proper mitochondrial protein folding in β cells.
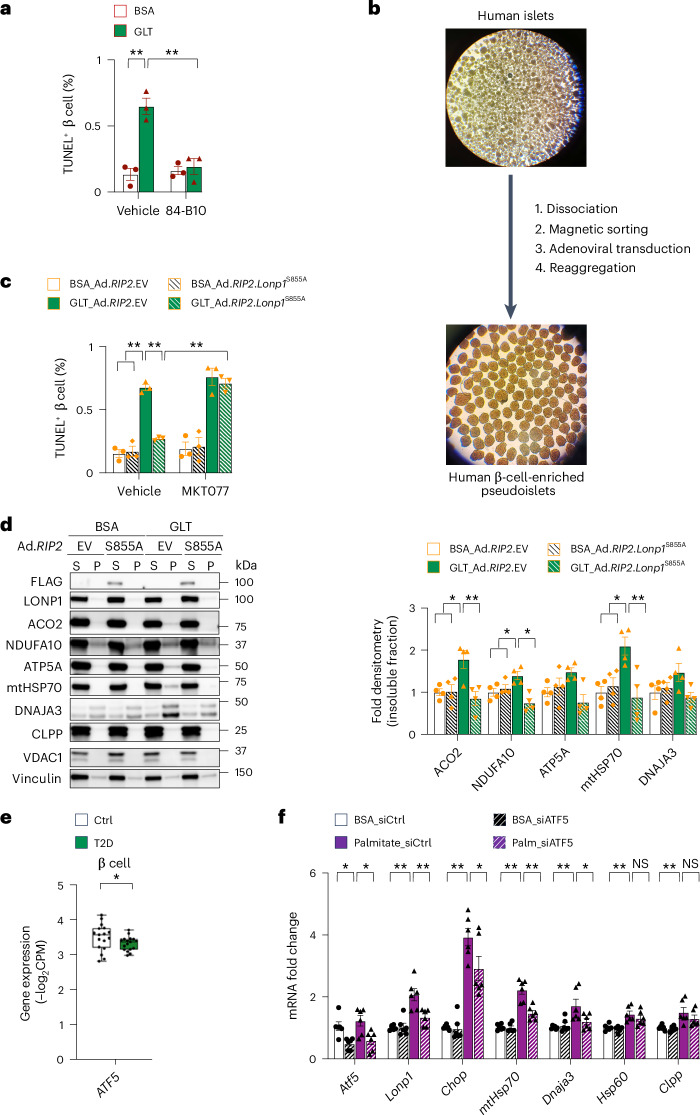
Finally, to explore the relevance of LONP1 and its mechanism of action in a model of T2D in human β cells, we assessed β cell survival in human islets subjected to GLT. Notably, GLT elicits β cell toxicity, the UPR and mitochondrial structural and functional defects64, yet a role for GLT to induce mitochondrial protein misfolding, similar to our observations in the human islets of T2D donors (Fig. 1), has not previously been assessed. Thus, we first tested if the recently described 3-phenylglutaric acid derivative 84-B10, which promotes LONP1 activity60,65, could ameliorate β cell cytotoxicity after GLT. Similar to a previous report60, we confirmed that 84-B10 enhanced LONP1-mediated turnover of recombinant TFAM in vitro (Extended Data Fig. 10d). Importantly, we observed that 84-B10 rescued GLT-induced β cell apoptosis in human islets (Fig. 7a).
Fig. 7. LONP1 promotes human β cell survival and is transcriptionally regulated by ATF5 after GLT.
a, Quantification of TUNEL staining for β cell apoptosis in human islets after exposure to BSA or GLT together with DMSO or 40 μM of the LONP1 activator 84-B10 for 48 h. n = 3 independent human islet donors per group. b, Schematic diagram illustrating the generation of human β-cell-enriched pseudoislets. c, Quantification of TUNEL staining for β cell apoptosis in human β-cell-enriched pseudoislets 7 days after adenoviral transduction with RIP2-driven EV (Ad.RIP2.EV) or the protease-deficient Lonp1S855A mutant (Ad.RIP2.Lonp1S855A), followed by exposure to BSA or GLT for the final 48 h, and DMSO or 1 μM MKT077 for the final 24 h. n = 3 independent human islet donors per group. d, Representative immunoblots (left) of mitochondrial proteins from the soluble and insoluble fractions of human β-cell-enriched pseudoislets 8 days after Ad.RIP2.EV or Ad.RIP2.Lonp1S855A transduction, exposed to BSA or GLT for the final 72 h. Quantification of mitochondrial insoluble proteins using densitometry (normalized to vinculin) is shown in the graph on the right. VDAC1 serves as a soluble mitochondrial protein loading control. Vinculin serves as a loading control for both soluble and insoluble fractions. n = 4 independent islet donors per group. e, Pseudobulk gene expression of ATF5 from the β cells of human islet donors with or without T2D using scRNA-seq. The box plots present the minimum, first quartile, median, third quartile, maximum and interquartile range. n = 17 donors without T2D, n = 17 donors with T2D. f, RT–qPCR of Atf5 and markers of the UPRmt from RNA isolated from Min6 β cells 72 h after transfection with siATF5 or siCtrl, and exposure to 0.5 mM palmitate or BSA control for the final 48 h. n = 6 per group. All data are presented as the mean ± s.e.m. a,c,d,f, *P < 0.05 and **P < 0.01 were determined using a one-way ANOVA followed by Tukey’s multiple comparisons test. e, *P < 0.05 was determined using both an unpaired, two-tailed Student’s t-test and FDR < 5% for multiple testing correction.
To determine if the protease or chaperone activity of LONP1 was necessary to promote human β cell survival and mitochondrial protein folding, we next generated human β-cell-enriched pseudoislets after magnetic sorting for the β cell marker NTPDase3 for better visualization of β cell proteins in subsequent biochemical assays (Fig. 7b). Indeed, we confirmed that magnetic sorting for NTPDase3 yielded ~90–95% insulin+ β cells (Extended Data Fig. 10e)66. Human β cells were then transduced with Ad.RIP2.Lonp1S855A (or Ad.RIP2.EV Ctrl) before pseudoislet generation. Importantly, overexpression of Lonp1S855A rescued GLT-induced β cell apoptosis (Fig. 7c). Similar to the observations with mouse islets above, the effects of LONP1 on the rescue of β cell survival after GLT were dependent on LONP1-mtHSP70 activity, as the benefits of Lonp1S855A overexpression were abrogated in the presence of mtHSP70 inhibition by MKT077 exposure (Fig. 7c). Furthermore, we observed that GLT exposure elicited a signature of mitochondrial protein misfolding in human β cells, which was reversed after overexpression of the Lonp1S855A mutant (Fig. 7d). Taken together, these findings indicate that LONP1 acts together with the chaperone mtHSP70 to regulate β cell survival.
ATF5 regulates the expression of LONP1 and the UPRmt in β cells after lipotoxicity
Given our observation of reduced LONP1 mRNA expression in the β cells of human islet donors with T2D (Fig. 2a), we evaluated transcriptional regulators of LONP1 to ascertain a possible mechanism underlying lower LONP1 expression in T2D. Several upstream transcriptional regulators of LONP1 have been reported, including GABPA67, NF-κB67, STAR68, HIF1α69 and ATF5 (ref. 70). Thus, we measured the expression of these transcriptional regulators in the β cells of human islet donors, finding a significant reduction only in ATF5 expression in human β cells of donors with T2D (Fig. 7e and Extended Data Fig. 10f). Interestingly, ATF5 binds to the promoter of LONP1 and directly regulates LONP1 expression70. ATF5 also regulates the UPRmt in HEK 293T cells and maintains β cell survival after cellular stress70,71.
To evaluate the importance of ATF5 in the regulation of β cell Lonp1 expression, we first exposed Min6 β cells to palmitate for 48 h to induce lipotoxicity, noting that palmitate exposure promoted the expression of Lonp1 and the UPRmt (Fig. 7f). We found that induction of LonP1 and the UPRmt was dependent on Atf5, as small interfering RNA-mediated knockdown of Atf5 reduced the expression of Lonp1 and the UPRmt elicited by palmitate exposure (Fig. 7f). Thus, these studies suggest that ATF5 regulates the expression of Lonp1 and the UPRmt in β cells after lipotoxic metabolic stress. These studies also lead us to speculate that reduced Lonp1 expression and the lack of the induction of the UPRmt in β cells in T2D, despite the presence of mitochondrial protein aggregates, could be due in part to reductions in ATF5.
Discussion
In this study, we identified impaired mitochondrial proteostasis in T2D islets and elucidated the impact of mitochondrial protein misfolding on β cell survival and glycaemic control. We found that the β cells of donors with T2D exhibited reduced expression of the matrix protease LONP1, and that loss of LONP1 elicits an accumulation of misfolded mitochondrial proteins that subsequently yields bioenergetic defects and oxidative stress. Intriguingly, LONP1 interacts with the chaperone mtHSP70 and functions in a protease-independent manner together with mtHSP70 to promote mitochondrial protein folding and rescue human β cell survival after GLT to model the metabolic stress of T2D. Furthermore, impairments in the expression of LONP1 and transcriptional activation of the UPRmt in the β cells of donors with T2D in response to mitochondrial protein aggregates may be caused by reductions in ATF5 expression. Together, our studies establish mitochondrial proteostasis as a crucial regulator of β cell survival and provide insights into mechanisms of β cell failure in T2D.
To understand the extent of impaired proteostasis in T2D, we applied a commonly used biochemical approach to quantitative islet proteomics by identifying changes in protein solubility. We were surprised to observe a robust enrichment in insoluble proteins within the mitochondrial matrix in the islets of donors with T2D. Furthermore, we unexpectedly found that the islets of donors with T2D had a proteomic signature favouring protein misfolding in the mitochondria to an increased extent than in the ER, positioning mitochondrial proteostasis as an important pathway in the pathology of islets in T2D. A limitation of our proteomics approach includes the use of filter-aided processing to remove the detergents needed to solubilize the insoluble fraction of human islets for TMT–MS, while this approach was not necessary in the soluble fraction, thus raising a potential for confounding effects when comparing results from distinct sample processing methods. This limitation also reinforces the need to confirm proteomics results using independent approaches where filter-aided processing was not used, which we did perform using immunoblotting. Additionally, our proteomics results were based on a limited number of human islet samples because of the paucity of donors with T2D, including a donor with T2D who developed prominent hyperglycaemia and weight loss in the context of medication non-adherence in the 5 months before death. However, recent rigorous quantitative proteomics studies on islets from donors with T2D support the observation of defective mitochondrial metabolism in T2D14,72. Together with these studies, our work supports tailoring the use of future discovery studies in T2D to discern organelle-specific or compartment-specific events that transpire to yield β cell dysfunction. Future studies may also be expanded to include spatial single-cell technologies in T2D, such as co-detection by indexing and imaging mass cytometry, to leverage high-throughput visualization of islet cell organization and composition towards the evaluation of organelle-specific events73,74.
Models of LONP1 deficiency display many mitochondrial defects, including the accumulation of misfolded mitochondrial protein aggregates, increases in mtROS, alterations in the ETC–OXPHOS system and reductions of mitochondrial mass28–30,54. We observed many of these effects in β cells bearing LONP1 deficiency and endeavoured to identify an initiating cause of β cell dysfunction and apoptosis. Indeed, we observed that oxidative damage was a slight contributor to loss of β cell mass and hyperglycaemia in β-Lonp1 KO mice; however, overexpression of a mitochondrial-targeted catalase was insufficient to remedy mitochondrial protein misfolding and only delayed, but did not prevent, β cell failure. We found reductions in glucose-stimulated bioenergetics and cytosolic [Ca2+] in β-Lonp1 KO mice, while mitochondrial [Ca2+] was surprisingly unchanged; mitochondrial [Ca2+] in β cells is often considered to be dependent on both of these aforementioned contributors75. Maintenance of mitochondrial [Ca2+], despite a decline in cytosolic [Ca2+], after LONP1 deficiency may be a consequence of both enhanced stability of the mitochondrial calcium uniporter, which regulates mitochondrial Ca2+ uptake, as well as loss of function of the Na+/Ca2+ exchanger, which regulates mitochondrial Ca2+ efflux, both of which have been reported previously76,77. Thus, understanding the properties of LONP1-deficient β cells to preserve mitochondrial [Ca2+] despite a decline in cytosolic [Ca2+] will be intriguing for future work. We also observed the induction of mitochondrial protein misfolding, including key ETC–OXPHOS proteins, which was associated with reduced levels of fully assembled ETC complexes that preceded elevations in ROS and reductions in mitochondrial mass. Interestingly, LONP1 deficiency also elicits autophagy and mitophagy as a compensatory measure to clear mitochondria bearing misfolded proteins, which will be of interest for future investigations given the importance of autophagy and mitophagy in β cells25,26,78. We observed that LONP1 interacts with and functions in a protease-independent manner together with the chaperone mtHSP70 to regulate β cell survival and prevent mitochondrial protein misfolding. These results suggest that in β cells, prevention of mitochondrial protein misfolding by LONP1 may be vital to avert a cascade of additional mitochondrial defects.
While LONP1 is not a known risk gene for T2D, our studies suggest that connections between LONP1, mitochondrial protein misfolding and β cell viability are of value to the understanding of T2D. A question raised from our work is not only the origin of reduced β cell LONP1 expression in T2D, but also the additional failure to upregulate LONP1, mtHSP70 or the UPRmt in response to mitochondrial protein misfolding, a phenomenon normally expected to occur in β cells after exposure to lipotoxicity, which we observed in this study and also reported previously by others24,31. Our studies suggest that reduced LONP1 expression and the failed transcriptional response to mitochondrial protein misfolding in T2D could be related to reductions in the expression of ATF5, a key regulator of the mammalian UPRmt. Impairments in ATF5 could also offer a target for understanding β cell dysfunction in T2D and open future questions as to the aetiology of reduced ATF5 expression itself in T2D β cells. Relatedly, understanding possible posttranscriptional regulators of LONP1 in β cells may also be vital in T2D. In other cell types, LONP1 is posttranscriptionally regulated by cap-independent translation in models of ageing79, oxidative repression of its activity in models of heart failure80, by sirtuin-3-dependent deacetylation81 and by autoregulatory mechanisms82, which may also warrant future exploration in T2D. Of course, these defects may be related to polymorphisms in other nuclear-encoded mitochondrial genes associated with T2D15–21. Furthermore, the induction of mitochondrial protein misfolding by exposure to GLT may suggest that acute diabetogenic stimuli could begin to overwhelm the endogenous mitochondrial proteostasis machinery in β cells. Thus, studies focused on the intrinsic capacity of β cells to balance mitochondrial proteostasis when facing metabolic stress will be of interest for future work. Assessment of the temporal progression of β cell mitochondrial proteostatic defects towards the development of T2D is not feasible in human donors with T2D but would be of value in mouse models of T2D. While some mouse models of T2D, such as the leptin-receptor-deficient db/db BKS strain, develop some, although not all, of the mitochondrial quality control defects found in T2D10–12,49, it is unknown whether db/db mice or other models of T2D develop mitochondrial protein misfolding. Our observation of improvements in mitochondrial protein folding and cell survival of human β cells after GLT by LONP1 overexpression or pharmacological activation could also inspire future therapeutic targets related to LONP1.
T2D is a complex polygenic and multi-organ disease driven by β cell dysfunction and complicated by extrinsic metabolic and chronological stress whose aetiopathological mechanisms remain vexing. Our studies have identified a convergence of two key β cell defects of impaired proteostasis and mitochondrial dysfunction in T2D caused by mitochondrial protein misfolding. Given the promising effects to improve β cell survival using pharmacological chaperones to offset ER protein misfolding, as well as proteases targeting IAPP, our results suggest that future approaches targeting multiple proteostatic pathways, including mitochondrial proteostasis, might together be beneficial to counter the many proteotoxic insults observed in T2D83,84. The convergence of impaired proteostasis and mitochondrial dysfunction in T2D because of mitochondrial protein misfolding, a previously unappreciated concept in β cells in the context of diabetes, also fits within the expanding context and understanding of ageing and ageing-related diseases, where impaired mitochondrial proteostasis is well known to occur1. Indeed, a renewed focus on T2D through the underappreciated lens of shared mechanisms of ageing-related diseases could provide an attractive method to approach T2D in the future. Interestingly, activation of mitochondrial proteases and chaperones in the absence of mitochondrial damage showed promise in extending the lifespan of Caenorhabditis elegans85. Thus, it is intriguing to speculate if the activation of mitochondrial proteases and chaperones to improve mitochondrial proteostasis could also improve β cell metabolic efficiency and viability to treat or prevent T2D.
Methods
Genetically modified mouse lines
All mice were maintained in accordance with the University of Michigan’s Institutional Animal Care and Use Committee under specific pathogen-free conditions. Up to five mice were housed per cage on a standard 12-h light–12-h dark cycle at room temperature and 30–70% humidity, with ad libitum access to food and water. All mice were fed PicoLab Laboratory Rodent Diet 5L0D (LabDiet) containing 28% protein, 13% fat and 57% carbohydrates. All experiments were performed with both male and female mice; result data from both sexes were combined in all experimental groups. Lonp1loxP/loxP tm1a mice possessing loxP sites flanking exon 2 of the Lonp1 gene were obtained from the European Mutant Mouse Archive. These animals were initially crossed to the ACTB-FLPe line to achieve deletion of the Frt-flanked neomycin cassette86 before the generation of tissue-specific KO models. To generate β-cell-specific deletion, floxed models were crossed to Ins1Cre mice from The Jackson Laboratory (strain no. 026801). Ins1Cre alone and floxed-only controls (Lonp1loxP/loxP and LonP1loxP/+ mice) were phenotypically indistinguishable from each other and were combined as Ctrls. Ins1Cre alone mice were previously shown to be phenotypically indistinguishable from wild-type (WT) C57BL/6N Ctrls in previous reports from our group and others10,49,87–89. For inducible β-cell-specific deletion, MIP1-CreERT mice (strain no. 024709, The Jackson Laboratory) were used or crossed to Lonp1loxP/loxP mice to generate iβ-Lonp1 KO mice. Eight-week-old MIP1-CreERT; Lonp1loxP/loxP mice and MIP1-CreERT mice were administered 100 mg kg day−1 TMX (cat. no. T5648, Sigma-Aldrich) dissolved in sterile filtered sunflower seed oil (Sigma-Aldrich) by intraperitoneal injection for 5 consecutive days. Filtered sunflower seed oil was administered into MIP1-CreERT; Lonp1loxP/loxP mice as an additional control group. Floxed stop mCAT overexpression mice (strain no. 030712, The Jackson Laboratory) were also bred to generate mCAT; β-Lonp1 KO mice. Ins1Cre alone and mCAT; Ins1Cre littermates were used as Ctrls for these studies.
Human islet samples
All human samples were procured following islet isolation from the pancreas of donors with or without T2D by the Integrated Islet Distribution Program or Prodo Laboratories, which receive samples from de-identified organ donors for research. Studies were approved by the University of Michigan Institutional Review Board. Human primary islets were cultured at 37 °C with 5% CO2 in PIM(S) medium (Prodo Labs) supplemented with 10% FCS (GeminiBio), 100 U ml−1 penicillin/streptomycin (Gibco), 100 U ml−1 antibiotic/antimycotic (Gibco) and 1 mM PIM(G) (Prodo Labs). Islets were obtained from male and female donors; de-identified donor information is provided in Supplementary Tables 1–3. Drug treatments used for human islet cultures included 1 μM CDDO (Cayman Chemical), 1 μg ml−1 tunicamycin (Cayman Chemical) or 5 mM NAC for 24 h, respectively, as well as the respective vehicle Ctrls (DMSO or PBS). Neutral-buffered, formalin-fixed, paraffin-embedded human pancreas sections were procured from de-identified donors with or without T2D from the Integrated Islet Distribution Program.
Mouse primary islet isolation and culture
Mouse primary islets were isolated by perfusing pancreata with a 1 mg ml−1 solution of Collagenase P (Sigma-Aldrich) in 1× HBSS into the pancreatic duct. After excision of the pancreas, pancreata were incubated at 37 °C for 13 min; Collagenase P was deactivated by adding 1× HBSS + 10% adult bovine serum (Quench buffer). Pancreata were dissociated mechanically using vigorous shaking for 30 s and the resulting cell suspension was passed through a 70-μM cell strainer (Thermo Fisher Scientific). Cells were centrifuged at 200g for 2 min, the pellet was resuspended in 20 ml Quench buffer and gently vortexed to thoroughly mix. Cells were again centrifuged at 200g for 1 min. This wash step was repeated once more. After the washes, the cell pellet was resuspended in 5 ml Histopaque (Sigma-Aldrich) with gentle vortexing. An additional 5 ml Histopaque was layered on the cell suspension; finally, 10 ml Quench buffer was gently layered on top. Cells were spun at 800g for 30 min at 10 °C, with the brake off. The entire Histopaque gradient was pipetted off and passed through an inverted 70-μM filter to trap the islets cells. Islets were washed twice with 10 ml Quench buffer and once with complete islet medium (Roswell Park Memorial Institute 1640 medium supplemented with 100 U ml−1 penicillin/streptomycin, 10% FCS, 1 mM HEPES, 2 mM l-glutamine, 100 U m−1 antibiotic/antimycotic and 10 mM sodium pyruvate). The filter was inverted into a sterile Petri dish and cells were washed into the dish with 4.5 ml complete islet medium.
Cell culture and transfections
Min6 β cells (a gift from D. Stoffers, University of Pennsylvania) between passages 28 and 35 were cultured with 25 mM glucose (Invitrogen) supplemented with 10% heat-inactivated FCS, 100 U ml−1 penicillin/streptomycin and 10 mM sodium pyruvate. For the knockdown studies, Min6 cells were transfected with an Amaxa nucleofector (Cell Line Nucleofector Kit V, Program G-016, Lonza Bioscience) or Lipofectamine RNAiMAX (Invitrogen) by delivering On-TARGET Plus siRNA SMART pools (Horizon Discovery) against Lonp1 (5′-UCGGAGACAAGUUGCGAAU-3′, 5′-AGAAAGGACUACUCGGAUU-3′, 5′-GAAAGAGAGUGCCCGCAUA-3′, 5′-CGAGAAUACUUCCGUUUCA-3′), Atf5 (5′-ACGUCUGUCUCCAGCGUCA-3′, 5′-CGGGAGAGUCAGUACGUGA-3′, 5′-GCUCUCAGGUACCGCCAGA-3′, 5′-GGGCUGGCUCGUAGACUAU-3′) or non-targeting control (5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′, 5′-UGGUUUACAUGUUUUCCUA-3′) for 72 h for the downstream studies. For the overexpression studies, Min6 cells were transfected with Lipofectamine 3000 (Invitrogen) with previously described pQCXIP plasmids expressing FLAG-epitope-tagged human WT LONP1, protease-deficient Lonp1S855A mutant or EV for 72 h29.
Pseudoislets
Islets from 3–4-week-old Ctrl and β-Lonp1 KO mice were hand-picked and then dispersed with trypsin (Thermo Fisher Scientific). Islet cells were counted and incubated with adenovirus expressing an EV Ctrl (Ad.RIP2.EV) or a protease-deficient Lonp1S855A mutant under control of the rat insulin 2 promoter (Ad.RIP2.Lonp1S855A) for 2 h at a multiplicity of infection (MOI) of 250. Cells were then seeded at 2,000 cells per well in CellCarrier spheroid ultra-low attachment microplates (PerkinElmer) in enriched pseudoislet medium as described previously49. Cells were allowed to reaggregate for 7 days before being collected for the downstream studies. For the in vitro Lonp1 deletion in adult mouse pseudoislets, islets from 7–8-week-old MIP1-CreERT; Lonp1loxP/loxP mice were dispersed and transduced with Ad.RIP2. EV and Ad.RIP2.Lonp1S855A) for 2 h at an MOI of 250, and then seeded at 2,000 cells per well in enriched pseudoislet medium supplemented with 2 μM 4-OHT (Sigma-Aldrich) or vehicle control (0.1% ethanol) to induce recombination, followed by biochemical assays.
To generate β-cell-enriched human pseudoislets, primary human islets were first dissociated with TrypLE Express Enzyme (Gibco) before incubation with a mouse monoclonal anti-human NTPDase3 antibody (ectopeptidases) for 30 min at 4 °C. Dissociated human islets were then washed in a magnetic-activated cell sorting buffer containing 1× sterile PBS (Corning), 0.5% FCS and 1% penicillin/streptomycin solution and then pelleted before resuspension in magnetic-activated cell sorting buffer containing anti-mouse IgG2a+b MicroBeads (Miltenyi Biotec) and incubation for 15 min at 4 °C. NTPDase3+ β cells were then purified using LS columns (Miltenyi Biotec), followed by counting and incubation with Ad.RIP2.EV or Ad.RIP2.Lonp1S855A for 2 h at an MOI of 50, and then seeded at 2,000 cells per well in enriched pseudoislet medium as above. Cells were allowed to reaggregate for 5 days before incubation with GLT medium containing 25 mM glucose + 0.5 mM palmitate (conjugated to BSA as described previously90) or control medium containing 5 mM glucose + BSA alone for 48–72 h before the downstream studies. Drug treatments used for pseudoislet studies included 40 μM 84-B10 (MedChemExpress) for 48 h or 1 μM MKT077 (Cayman Chemical) for 24 h, respectively, as well as the respective vehicle controls (DMSO).
IPGTT
Mice were fasted for 6 h. Fasting blood glucose measurements were taken via tail nick (Bayer Contour glucometer) before an intraperitoneal injection of 1 g kg−1 glucose was administered. Blood glucose measurements were then taken at 15, 30, 60 and 120 min. After the test, mice were returned to the housing cages with ad libitum access to food and water.
In vivo glucose-stimulated insulin release
Mice were fasted for 6 h. Fasting blood glucose was measured via tail nick with a glucometer (Bayer Contour); a 20-μl blood sample was collected using capillary tubes (Thermo Fisher Scientific) and stored on ice. Mice were injected with 3 g kg−1 glucose, and blood glucose and blood samples were taken after 3 min. Blood samples were ejected from the capillary tubes into 1.5-ml tubes, spun at 10,000g, 4 °C for 10 min, and serum was aliquoted to new 1.5-ml tubes. Serum insulin levels were measured using ELISA (ALPCO Diagnostics). After the test, mice were returned to the housing cages with ad libitum access to food and water.
Intraperitoneal insulin tolerance tests
Mice were fasted for 6 h. Fasting blood glucose was measured via tail nick with a glucometer (Bayer Contour). Mice were injected with 0.8 U kg−1 insulin (Humulin R, Eli Lilly and Company) and blood glucose was measured at 15, 30 and 60 min. After the test, mice were returned to the housing cages with ad libitum access to food and water.
Islet respirometry
Islet respirometry was performed using the BaroFuse instrument (EnTox Sciences) as described previously91. The set-up of the BaroFuse perfusion instrument involved four steps: (1) preheating the system followed by filling the medium reservoirs with pre-equilibrated medium (1× Krebs–Ringer buffer, HEPES-buffered, with 3 mM glucose); (2) inserting the tissue chamber assemblies into the perfusion module and securing it in place on top of the medium reservoir to create a gas tight seal; (3) purging the headspace of the medium reservoirs with the desired gas composition (typically 5% CO2, 21% O2, balance N2) and allow the gas in the headspace to equilibrate with the perfusate in the reservoir; and (4) closing the purge port to allow the flow to fill up the tissue chambers. When the tissue chambers had almost filled, islets were loaded into six of the eight chambers and two were left empty to be used as an inflow reference. Two hundred hand-picked islets were transferred into the tissue chamber (ID = 1.5 mm) with a P-200 pipette. Once the islets were loaded, the magnetic stirrers, O2 detector and flow rate monitor were started, and the system and islets were allowed to equilibrate and establish a stable baseline for 90–120 min. Subsequently, the test compounds (final concentrations of 20 mM glucose, 10 μM oligomycin and 3 mM KCN) were injected at precise times. Data are presented as the fractional change from baseline (at 3-mM glucose-containing medium), the OCR per sample relative to the minimum OCR per sample determined after exposure to 3 mM KCN for baseline normalization using the BaroFuse Data Processing Package Software (EnTox Sciences).
Biochemical assessment of protein solubility
Human and mouse islets were lysed with Triton X-100 buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) supplemented with protease and phosphatase inhibitors (Protease Inhibitor Set III and Phosphatase Inhibitor Set II, Calbiochem), incubated on ice for 30 min and then centrifuged at 21,000g for 10 min at 4 °C to separate into a supernatant (Triton X-100 soluble fraction) and pellet (Triton X-100 insoluble fraction). Insoluble fractions were dissolved in 2% SDS containing a 50% volume of Triton X-100 buffer (with protease and phosphatase inhibitors), followed by immunoblotting or TMT labelling MS.
SDS–PAGE
Isolated mouse islets or human pseudoislets were homogenized in radio-immunoprecipitation assay buffer containing protease and phosphatase inhibitors. Samples were centrifuged at 200g for 5 min at 4 °C and the supernatant was used for the immunoblot analysis. Protein quantification was carried out using a Pierce Micro BCA Kit (Thermo Fisher Scientific). Protein lysates were prepared in Laemmli buffer with dithiothreitol and denatured at 70 °C for 10 min, or 37 °C for 30 min for OXPHOS analysis. Samples were then run on a 4–15% Tris-glycine protein gel (Bio-Rad Laboratories) at 150 V until separated. Samples were then transferred to a nitrocellulose membrane at 90 V for 90 min; membranes were blocked with 5% skimmed milk in 1× TBS + 0.1% Tween-20 for 1 h and incubated overnight at 4 °C with primary antisera, including LONP1 (1:1,000 dilution, cat. no. 15440-1-AP, Proteintech), GRP75 (mtHSP70, 1:1,000 dilution, cat. no. 14887-1-AP, Proteintech), HSP60 (1:1,000 dilution, sc-13115, Santa Cruz Biotechnology), DNAJA3 (1:1,000 dilution, cat. no. sc-18820, Santa Cruz Biotechnology), CLPP (1:1,000 dilution, cat. no. 15698-1-AP, Proteintech), aconitase 2 (1:1,000 dilution, cat. no. ab110321, Abcam), NDUFA10 (1:1,000 dilution, cat. no. sc376357, Santa Cruz Biotechnology), SDHA (1:1,000 dilution, cat. no. ab14715, Abcam), total OXPHOS rodent antibody cocktail (1:1,000 dilution, cat. no. ab110413, Abcam), total OXPHOS human antibody cocktail (1:1,000 dilution, cat. no. ab110411, Abcam), TOM20 (1:1,000 dilution, cat. no. 42406, Cell Signaling Technology), VDAC1 (1:1,000 dilution, cat. no. ab14734, Abcam), catalase (1:2,000 dilution, cat. no. 01-05-30000, Athens Research and Technology), phospho-γH2AX (1:1,000 dilution, cat. no. 05-636, Merck Millipore), FLAG (1:1,000 dilution, cat. no. F1804, Sigma-Aldrich), Twinkle (1:1,000 dilution, cat. no. 13435-1-AP, Proteintech), HMGCS2 (1:1,000 dilution, cat. no. A14244, ABclonal), TFAM (1:1,000 dilution, cat. no. ab119684, Abcam) and vinculin (1:1,000 dilution, cat. no. CP74, Merck Millipore). Membranes were then washed three times with 1× TBS + 0.1% Tween-20 and incubated with species-specific horseradish peroxidase-conjugated secondary antisera (Vector Laboratories).
BN-PAGE
BN-PAGE was performed similar to a previously published protocol as described in ref. 92. Briefly, mouse islets were lysed in native sample buffer (62.5 mM Tris-HCl, 10% glycerol and protease inhibitors, pH 7.0) containing 1% n-dodecyl-beta-d-maltoside (Sigma-Aldrich) and 1% digitonin (Sigma-Aldrich), incubated on ice for 30 min and centrifuged at 21,000g and 4 °C for 15 min. Equal quantities of protein lysates, determined using a Micro BCA Protein Assay Kit (Pierce) were added to 4–15% polyacrylamide gradient gels and supplemented with Coomassie Brilliant Blue (0.2% final concentration, Thermo Fisher Scientific) added to the supernatant. After electrophoresis, proteins were transferred to activated polyvinylidene fluoride membranes, followed by immunoblotting. Immunoblotting was performed using NDUFA9 (1:1,000 dilution, cat. no. ab14713, Abcam), SDHA (1:1,000 dilution, cat. no. ab14715, Abcam), UQCRC2 (1:1,000 dilution, cat. no. ab14745, Abcam), total OXPHOS Blue Native immunoblotting antibody cocktail (1:1,000 dilution, cat. no. ab110412, Abcam) and species-specific horseradish peroxidase-conjugated secondary antibodies (Vector Laboratories).
β cell mass analysis
The whole pancreas from each mouse was excised, weighed and fixed in 4% paraformaldehyde for 16 h at 4 °C. Samples were stored in 70% ethanol, at 4 °C, before being embedded in paraffin and sectioned. Three independent depths of sections, at least 50 μM apart, were dewaxed and rehydrated; antigen retrieval was carried out using 10 mM sodium citrate (pH 6.0) in a microwave for 10 min. Sections were washed twice with 1× PBS, blocked for 1 h at room temperature with 5% donkey serum in PBS with Tween-20 (PBT) (1× PBS, 0.1% Triton X-100, 1% BSA). Sections were then incubated in the following primary antisera overnight at 4 °C in PBT: guinea pig anti-insulin (cat. no. PA1-26938, Thermo Fisher Scientific). Sections were then washed twice with PBS and incubated for 2 h at room temperature with species-specific Cy2-conjugated secondary antibody. Nuclear labelling was performed using DAPI (Molecular Probes). Sections were scanned using an Olympus IX-81 microscope at ×10 magnification, with image stitching for quantification (Olympus). β cell mass quantification (estimated as the total insulin+ area divided by the total pancreatic area and multiplied by pancreatic weight) was performed on stitched images of complete pancreatic sections from three independent regions.
Immunofluorescence imaging
The whole pancreas from each mouse was excised, weighed and fixed in 4% paraformaldehyde for 16 h at 4 °C. Samples were stored in 70% ethanol at 4 °C before being embedded in paraffin and sectioned. After dewaxing and rehydration, antigen retrieval was performed in 10 mM citric acid (pH 6.0) or HistoVT buffer (Nacalai Tesque) as described previously49. Samples were then blocked in PBT containing 5% donkey serum or Blocking One solution (Nacalai Tesque) for 1 h at room temperature. For human pancreas sections, samples were dewaxed and rehydrated before antigen retrieval in 10 mM citric acid (pH 6.0) as noted earlier. Samples were then blocked in PBT containing 5% donkey serum for 1 h at room temperature. Mouse or human pancreatic sections were stained with the following primary antibodies: insulin (cat. no. PA1-26938, Thermo Fisher Scientific, or cat. no. 20-IP35, Fitzgerald); Ki-67 (cat. no. ab16667, Abcam), PDX1 (cat. no. ab47383, Abcam); ALDH1A3 (cat. no. NBP2-15339, Novus Biologicals); urocortin 3 (cat. no. PBL7218, Salk, a gift from the laboratory of W. Vale); GRP75 (mtHSP70, cat. no. 14887-1-AP, Proteintech); SDHA (cat. no. ab14715, Abcam); C-peptide (cat. no. GN-1D4, Developmental Studies Hybridoma Bank, deposited by O. D. Madsen, Hagedorn Research Institute); total OXPHOS human antibody cocktail (cat. no. ab110411, Abcam); and species-specific Cy2, Cy3 and Cy5 conjugated secondary antibodies (Jackson ImmunoResearch) or PROTEOSTAT Aggresome Detection Kit (cat. no. ENZ-51035, Enzo Life Sciences). Nuclear labelling was performed using DAPI. High-magnification images were captured under oil immersion on an Olympus IX-81 microscope.
Imaging of mitochondrial morphology in mouse β cells
Images of pancreas sections stained with SDHA and insulin (to identify β cells) were captured with an IX-81 microscope using an ORCA Flash4 complementary metal oxide semiconductor (CMOS) digital camera (Hamamatsu). Immunostained pancreatic sections were captured with Z-stack images and subjected to deconvolution (cellSens, Olympus). Mitochondrial morphology was visualized using 3D rendering generated with the Imaris imaging software (Bitplane). Quantitative 3D assessments of mitochondrial morphology and network were performed on ImageJ using the Mitochondria Analyzer plug-in93. Colocalization analyses were performed on Z-stack images of immunostained, dissociated islet cells using the Coloc2 plug-in in ImageJ.
qPCR of complementary and mitochondrial DNA content
DNA samples were isolated from mouse islets with the Blood & Tissue DNeasy Kit (QIAGEN). RNA samples were isolated from mouse islets or Min6 β cells with the MicroElute Total RNA Kit (Omega Bio-Tek) or TRIzol reagent (Invitrogen), respectively. DNase-treated RNA (Ambion DNA-free Kit) was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) as described previously94. qPCR was performed with SYBR-based detection (Universal SYBR Green Supermix, Bio-Rad Laboratories) using primers for mt9/11 for mtDNA (5′-GAGCATCTTATCCACGCTTCC-3′ and 5′- GGTGGTACTCCCGCTGTAAA-3′) and Ndufv1 (5′- CTTCCCCACTGGCCTCAAG-3′ and 5′-CCAAAACCCAGTGATCCAGC-3′) for nuclear DNA. For gene expression analysis of complementary DNA, primers included Atf5 (5′-CTGGGCTGGCTCGTAGACTAT-3′ and 5′-ATCAGAGAAGCCGTCACCTGC-3′), Lonp1 (5′-AAATCAAGGGGCACAGGCGTA-3′ and 5′-TTGGTAGCCTCGGCCAATCTT-3′), Chop (5′- CCTAGCTTGGCTGACAGAGG-3′ and 5′-CTGCTCCTTCTCCTTCATGC-3′), Grp75 (5′-AGGGCAAACAAGCAAAGGTCC-3′ and 5′-TGGTGACAGCTTGCCGTTTTG-3′), Dnaja3 (5′-AGAACCATGGATAGCTCCGCA-3′ and 5′-TCCAGTTGACCGCTTTCCTCA-3′), Hsp60 (5′-ACAATGGGGCCAAAGGGAAGA-3′ and 5′-GACTTTGCAACAGTGACCCCA-3′), Hsp10 (5′- GACTTTGCAACAGTGACCCCA-3′ and 5′- CTTTGGTGCCTCCATATTCTGGG-3′), Clpp (5′- GAACTGCGACGCGAGCTTTC-3′ and 5′-CACACTGTCGTCAATCGGGC-3′) and Hprt (5’-GGCCAGACTTTGTTGGATTTG-3′ and 5′- TGCGCTCATCTTAGGCTTTGT-3′).
TMT labelling MS-based quantitative proteomics
Human islets from donors with or without T2D were collected and lysed with Triton X-100 buffer containing protease and phosphatase inhibitors as described above for TMT labelling MS.
Protein digestion and TMT labelling
Depending on the solubilization buffer, proteins from insoluble fractions were initially processed using the FASP Protein Digestion Kit (cat. no. ab270519, Abcam); proteins from soluble fractions were processed using a standard TMT sample processing protocol (Thermo Fisher Scientific). Protein samples were digested with trypsin and Lys-C mix (1:25 protease: protein; Promega Corporation) at 37 °C with constant mixing using a thermomixer. The TMT 16-plex reagents were dissolved in 20 μl anhydrous acetonitrile and labelling was performed by transferring the entire digest to the TMT reagent vial and incubating at room temperature for 1 h. The reaction was quenched by adding 5 μl of 5% hydroxyl amine and further 15-min incubation. Labelled samples were mixed and dried using a vacufuge. Samples were randomly distributed across TMT channels to avoid potential channel-specific biases and technical variations among all experimental groups. The design of the sample labels for both insoluble and soluble fractions are provided in Supplementary Table 4. An offline fractionation of the combined sample (~200 μg) into eight fractions was performed using the High pH Reversed-Phase Peptide Fractionation Kit according to the manufacturer’s protocol (cat. no. 84868, Pierce). Fractions were dried and reconstituted in 9 μl of 0.1% formic acid and 2% acetonitrile in preparation for LC–MS2 analysis.
LC–multinotch MS3
To obtain superior quantitation accuracy, we used multinotch MS3, which minimizes the reporter ion ratio distortion resulting from fragmentation of co-isolated peptides during MS analysis. An Orbitrap Fusion (Thermo Fisher Scientific) and RSLC Ultimate 3000 nano-UPLC (Dionex) were used to acquire the data; 2 μl of the sample was resolved on a PepMap RSLC C18 column (75 μm × 50 cm internal diameter, Thermo Fisher Scientific) at a flow rate of 300 nl min−1 using a 0.1% formic acid and acetonitrile gradient system (2–22% acetonitrile in 150 min; 22–32% acetonitrile in 40 min; 20 min wash at 90% followed by 50 min re-equilibration) and directly sprayed onto the mass spectrometer using an EasySpray source (Thermo Fisher Scientific). The mass spectrometer was set to collect one MS1 scan (Orbitrap, 120 K resolution; AGC target 2 × 105; maximum IT = 100 ms) followed by data-dependent ‘Top Speed’ (3 s) MS2 scans (collision-induced dissociation; ion trap; NCE = 35; AGC 5 × 103; max IT = 100 ms). For multinotch MS3, the top ten precursors from each MS2 were fragmented using higher-energy collisional dissociation followed by Orbitrap analysis (NCE = 55; 60 K resolution; AGC = 5 × 104; maximum IT = 120 ms, 100–500 m/z scan range).
Data analysis
MS raw data were converted into open mzML format using the MSconvert tool from the ProteoWizard software suite and analysed using the FragPipe computational platform (https://fragpipe.nesvilab.org) using the established TMT16-MS3 workflow. MS2 spectra were searched using MSFragger (v.3.7) against the UniProt human reference proteome database (UP000005640, 20,389 entries, downloaded on 24th February 2023) appended with an equal number of decoy sequences. The search was restricted to tryptic peptides, allowing up to two missed cleavage sites. The precursor-ion mass tolerance was set to 20 ppm allowing C12 and C13 isotope errors—1/0/1/2/3. Mass calibration and parameter optimization were enabled. Carbamidomethylation of cysteine (57.02146 Da) and TMT labelling of lysine (304.2071 Da) were specified as fixed modifications; methionine oxidation (15.9949 Da), N-terminal protein acetylation (+42.0106 Da) and TMT labelling of the peptide N terminus (304.2071 Da) were specified as variable modification. The search results were further processed using Percolator to validate the reliability of peptide to spectrum matches (PSMs). The resulting files from Percolator were then converted to pep.xml format and processed to assemble peptides into proteins (protein inference) using ProteinProphet run via the Philosopher toolkit (v.5.0.0). The identified proteins were then filtered to keep only those that passed the 1% protein-level FDR using the Philosopher filter command. The PSM list was filtered using a sequential FDR strategy, keeping only those PSMs that passed the 1% PSM-level FDR and mapped to proteins that also passed the global 1% protein-level FDR. In addition, for all PSMs corresponding to a TMT-labelled peptide, reporter ion intensities were extracted from the high-quality MS3 spectra using Philosopher. The PSM output file from Philosopher was further processed using TMT-Integrator (v.4.0.4) to generate summary reports for protein and peptide quantification.
Downstream analysis
The quantification reports generated by TMT-Integrator were used for the downstream analysis. Differential expression analysis was conducted for all identified proteins under different treatments compared to DMSO, using the linear model provided by the limma package. Functional analysis of GO terms among all proteins was performed using the clusterProfiler package. To visualize the differential expression results, volcano plots were generated using the R package ggplot2. The log2 fold change was represented by limma’s moderated t-statistic on the x axis, and the −log10-transformed P value (−log10 P) on the y axis. MitoCarta3.0 targets were highlighted using green colour points. A log2 fold change greater than 0.1 or less than −0.1 was selected for the downstream analysis to capture small changes in larger protein complexes that may still be crucial for cellular function, to obtain a more comprehensive view of protein solubility for discovery purposes as a candidate-based approach and to ensure that changes in protein solubility that might be underestimated because of ratio compression of isobaric labelling are not missed95.
Adenoviral vectors and viral preparation
FLAG-tagged human Lonp1S855A was generated using PCR amplification of pQCXIP-Lonp1S855A (ref. 29) using PCR primers to add a FLAG epitope tag in-frame at the C terminus of LONP1. The PCR amplified fragment was subcloned into pCR-Blunt II-TOPO (Zero Blunt TOPO Cloning Kit, Invitrogen) for sequence validation using Sanger sequencing (Eurofins Scientific), followed by restriction digestion for KpnI and MfeI, and T4 ligation of fragments into a linearized pAd.RIP2 vector (provided by L. Satin, University of Michigan) for further adenovirus generation processing in the Vector Core at the University of Michigan. Briefly, pAd.RIP2.Lonp1S855A or pAd.RIP2 EV plasmids were cotransfected and recombined with cAd5 (E1–E3 genes deleted). Individual plaques were selected and viral lysates were screened for protein expression. Selected clones were then expanded in 911 cells and purified using CsCl gradient ultracentrifugation. Viral bands were collected and flushed through a Sepharose CL-4B (Cytiva) column. Eluted adenovirus was diluted in a sterile isosaline solution (10 mM Tris-HCl, 137 mM NaCl, 5 mM KCl, 1 mM MgCl2 and 10% glycerol (v/v)). Titres were determined by counting the plaque-forming units from 911 cells infected with serially diluted adenovirus and overlayed with 0.8% Noble agar in 1× EMEM + 5% FCS.
Fluorescence imaging of islet Ca2+ and mitochondrial membrane potential
Whole-islet fluorescence imaging
Mitochondrial Ca2+ uptake was measured in islets and β cells using adenovirus-mediated delivery of a mitochondrially targeted recombinant Ca2+ indicator, mito-R-GECO1 as described previously75. The Mito-R-GECO1 sequence was obtained from Addgene, and the adenoviral particles were generated by Vector Laboratories. Islets were infected within 1 h after isolation and incubated for 48 h. Of note, adenoviral tropism towards β cells ensured that Ca2+ changes were monitored largely in this cell type96. Whole islets were then loaded with the cytosolic Ca2+ indicator Cal-520 AM (2 µM; cat. no. ab171868, Abcam) in Krebs–Ringer bicarbonate buffer (140 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 24 mM NaHCO3, 1.5 mM CaCl2, 0.5 mM MgSO4, 10 mM HEPES and 3 mM d-glucose; pH 7.4) for 30 min. Images were captured at 1.64 Hz on a ZEISS Axio Observer Z1 spinning disc system equipped with a Fluor ×40/1.3 numerical aperture oil immersion objective at 490 nm excitation laser with a 561-nm emission filter, in low-glucose conditions (3 mM), followed by 11 mM glucose, 17 mM glucose and 20 mM KCl. Data were analysed using ImageJ.
Single-cell fluorescence imaging
For the mitochondrial membrane potential experiments with tetramethylrodamine methyl ester perchlorate (TMRM), islets were dissociated into single mouse islet cells and plated onto glass coverslips overnight at 37 °C. Cells were then loaded with 10 nM TMRM in Krebs–Ringer bicarbonate buffer at 3 mM glucose (pH 7.4) for 30 min and re-equilibrated with 2 nM TMRM for 10 min before imaging. TMRM (2 nM) was present throughout; images were taken in low-glucose conditions (3 mM), high-glucose conditions (17 mM) and after exposure to the uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (1 µM). The imaging experiments were captured as above with excitation at 561 nm, with an excitation laser with a 610-nm emission filter. Traces represent the mean normalized fluorescence intensity over time (F/Fmin), where Fmin is the mean fluorescence recorded at the end of the application of FCCP. Δψm was measured as the difference in mean fluorescence intensity measured at the peak after high-glucose administration (between 9.75 and 10.25 min) and the minimum prior glucose administration (between 2.5 and 3 min), and at the minimum after FCCP administration (between 13 and 13.5 min) and before FCCP administration (between 12.5 and 13 min).
Co-immunoprecipitation
Min6 β cells were lysed as described previously97, followed by shearing by passage through 21-gauge needle multiple times while on ice. Briefly, lysates were clarified using centrifugation, then pre-cleared with magnetic protein A/G beads (Thermo Fisher Scientific). Protein lysates were then incubated with magnetic protein A/G beads and control lgG (cat. no. 30000-0-AP, Proteintech) or LONP1 antibody overnight at 4 °C. Beads were washed three times in lysis buffer and conjugates were eluted in 4× loading buffer supplemented with 5 mM dithiothreitol (Thermo Fisher Scientific) at 70 °C for 10 min before SDS–PAGE.
ATP assay
Isolated islets from mice were cultured overnight at 37 °C. On the day of the experiment, 75 islets per group (low and high glucose for each genotype) were picked and preincubated in 1× Krebs–Ringer buffer (Thermo Fisher Scientific) for 30 min at 37 °C. For the high-glucose group, islets were incubated with 16.7 mM glucose for 5 min at 37 °C, while mouse islets for the low-glucose group remained in the Krebs–Ringer buffer with 2 mM glucose, and was then picked and collected in a microcentrifuge tube. Islets were then dissociated in trypsin, neutralized and then resuspended in 1× PBS. Single cells were pipetted into a 96-well white wall clear bottom plate. ATP concentrations were then measured using a Bioluminescence Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Luminescence was measured using a 5-s integration time. ATP concentrations were then normalized to total protein content (Micro BCA Kit, Pierce).
Enzyme activity assays
LONP1 activity was assessed via ATP-dependent degradation of the native mitochondrial protein substrate TFAM, similar to previously described approaches43,98. Briefly, purified recombinant human LONP1 (1-μM monomer) and 0.27 μM recombinant human TFAM (provided by C. Suzuki, Rutgers University) were suspended in a buffer consisting of 25 mM Tris, pH 7.9, 10 mM MgCl2 and 0.1 mg ml−1 BSA on ice, then preincubated for 30 min at 37 °C with 5 μM CDDO (Cayman Chemical), 40 μM 84-B10 (MedChemExpress) or DMSO control. An aliquot was taken representing time 0; the remaining sample was incubated at 37 °C for the indicated time after the addition of 2 mM ATP (Sigma-Aldrich). The samples were then subjected to SDS–PAGE. Aconitase activity was assessed in the lysates of Min6 β cells after exposure to 0.3 μM MKT077 or DMSO for 24 h with an Aconitase Assay Kit (Cayman Chemical) as per the manufacturer’s instructions.
Cell death and ROS assessments
Cytoplasmic histone-complexed DNA fragments were determined in islets using the Cell Death Detection ELISAPLUS (Roche), according to the manufacturer’s protocol. Cell pellets were analysed to detect apoptosis. ELISA absorbance was measured at 450 nm and normalized to total DNA content (SYBR Green, Invitrogen) using an absorbance and fluorescence microplate reader (BioTek). TUNEL was performed on mouse pancreatic tissue sections or transduced human islets (Apoptag In Situ Apoptosis Detection Kit, Merck Millipore) as per the manufacturer’s protocol. Islets were prepared for staining as described previously90. TUNEL-stained samples were counterstained with insulin antibodies. Images were captured with an IX-81 microscope using an ORCA Flash4 CMOS digital camera. TUNEL+ β cells were quantified using hand counting after image capture. ROS levels were measured using a cell-based fluorogenic assay kit (ROS/Superoxide Detection Assay Kit, Abcam) according to the manufacturer’s protocol, as described previously88. The gating strategy for the flow cytometry of ROS levels and the exclusion of cellular debris and doublets are shown in Supplementary Fig. 1.
Statistics
All data are presented as the mean ± s.e.m. Statistical analyses were performed in Prism 10.0 (GraphPad Software) using an unpaired, two-tailed Student’s t-test or a one-way or two-way ANOVA, followed by Tukey’s or Šidák’s post-hoc test for multiple comparisons as appropriate. A P < 0.05 was considered statistically significant. Data distribution was assumed to be normal, but this was not formally tested. Data collection and analysis were not performed blinded to the conditions of the experiments. No statistical methods were used to predetermine sample sizes; however, sample sizes were in line with those reported in previous publications10,49. Outlier tests (ROUT method99) were routinely performed in Prism and all results were included in the analysis unless designated by the ROUT method. No outliers were identified in the analysis of samples in this study. No data from experiments were excluded from the analysis of the groups in this study.
Materials availability
Lonp1loxP/loxP mice are available upon request from the lead author.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Tables 1–4 and Fig. 1.
Protein identifiers and quantitative measures in the insoluble fractions of human islets.
Protein identifiers and quantitative measures in the soluble fractions of human islets.
Source data
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data.
Statistical source data and unprocessed western blots.
Statistical source data.
Statistical source data.
Statistical source data and unprocessed immunoblots.
Statistical source data.
Statistical source data.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Acknowledgements
S.A.S. acknowledges support from Breakthrough T1D (grant nos. SRA-2023-1392 and SRA-2024-1586), the National Institutes of Health (NIH) (grant nos. R01 DK108921, R01 DK135032, R01 DK135268, R01 DK136671, R01 DK127270, U01 DK127747 and P30 DK020572), the Department of Veterans Affairs (no. I01 BX004444) and the Brehm and Anthony families. E.M.W. was supported by the NIH (grant no. K01 DK133533). J.L. acknowledges support from the American Diabetes Association (no. 1-25-PDF-126). M.L.S. acknowledges support from the ADA Pathway to Stop Diabetes Accelerator Award (no. 1-81-ACE-015) and the NIH (grant nos. R01 DK136671 and R01 DK118011). G.A.R. was supported by a Wellcome Trust Investigator Award (no. WT212625/Z/18/Z), a UK Research and Innovation Medical Research Council Programme grant (no. MR/R022259/1), Diabetes UK (BDA 16/0005485), an NIH-National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) project grant (no. R01DK135268), a Canadian Institutes of Health Research (CIHR)-Juvenile Diabetes Research Foundation (JDRF) team grant (nos. CIHR-IRSC TDP-186358 and JDRF 4-SRA-2023-1182-S-N), Centre Hospitalier de l’Université de Montréal start-up funds and an Innovation Canada John R. Evans Leader Award (no. CFI 42649). M.G. was supported by a Swiss National Science Foundation Postdoc mobility award no. P500PM_225305/1. We acknowledge the Microscopy, Imaging and Cellular Physiology Core and Islet Core of the University of Michigan DRC (no. P30 DK020572) for assistance with the imaging and pseudoislet studies, respectively. The proteomics studies were carried out in the Proteomics Resource Facility at the University of Michigan. Human pancreatic islets and other resources were provided by the NIDDK-funded Integrated Islet Distribution Program (Research Resource Identifier: SCR _014387) at City of Hope, NIH grant no. U24DK098085. We thank European Conditional Mouse Mutagenesis Program for providing the mutant LonP1 mouse line, INFRAFRONTIER-European Mutant Mouse Archive (www.infrafrontier.eu), and Helmholtz Zentrum München from which the mouse line was distributed (EM: 09070). We thank C. Suzuki, B. Kaufman, K. Claiborn, R. Noland and members of the Soleimanpour laboratory for helpful advice.
Extended data
Author contributions
J.L. conceived, designed and performed the experiments, interpreted the results, and drafted and reviewed the manuscript. Y.D. interpreted the results and reviewed the manuscript. M.G. designed and performed the experiments, and interpreted the results. J.Z. designed and performed the experiments, and interpreted the results. E.M.W. and V.S. designed and performed the experiments, interpreted the results and reviewed the manuscript. E.C.R., D.L.H., M.B.P., C.-S.S., K.B., E.M., S.N. and R.K. designed and performed the experiments, and interpreted the results. V.B. designed and performed the experiments, interpreted the results and reviewed the manuscript. A.I.N., M.L.S., D.C.C. and G.A.R. designed the studies, interpreted the results and edited and reviewed the manuscript. S.A.S. conceived and designed the studies, interpreted the results, and drafted, edited and reviewed the manuscript.
Peer review
Peer review information
Nature Metabolism thanks Cole Haynes, Jarrod Marto, Raghu Mirmira and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Revati Dewal, in collaboration with the Nature Metabolism team.
Data availability
The proteomics data have been deposited and are publicly available at the ProteomeXchange (PXD060404). Microscopy data will be shared by the lead contact upon reasonable request. Any additional information required to reanalyse the data reported in this paper is available from the lead author upon reasonable request. Source data are provided with this paper.
Competing interests
S.A.S. has received grant funding from Ono Pharmaceutical and is a consultant for Novo Nordisk. G.A.R. has received grant funding from Sun Pharmaceuticals and Les Laboratoires Servier and is a consultant for Sun Pharmaceuticals. A.I.N. receives royalties from the University of Michigan for the sale of MSFragger and IonQuant software licences to commercial entities. All licence transactions are managed by the University of Michigan Innovation Partnerships office; all proceeds are subject to university technology transfer policy. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s42255-025-01333-7.
Supplementary information
The online version contains supplementary material available at 10.1038/s42255-025-01333-7.
References
- 1.López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Kaushik, S. & Cuervo, A. M. Proteostasis and aging. Nat. Med.21, 1406–1415 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee, A., Morales-Scheihing, D., Butler, P. C. & Soto, C. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med.21, 439–449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzban, L., Park, K. & Verchere, C. B. Islet amyloid polypeptide and type 2 diabetes. Exp. Gerontol.38, 347–351 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Arunagiri, A. et al. Proinsulin misfolding is an early event in the progression to type 2 diabetes. eLife8, e44532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yong, J., Johnson, J. D., Arvan, P., Han, J. & Kaufman, R. J. Therapeutic opportunities for pancreatic β-cell ER stress in diabetes mellitus. Nat. Rev. Endocrinol.17, 455–467 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, C.-J. et al. High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress mediated β-cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes56, 2016–2027 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Prentki, M., Matschinsky, F. M. & Madiraju, S. R. Metabolic signaling in fuel-induced insulin secretion. Cell Metab.18, 162–185 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Xiong, S., Mu, T., Wang, G. & Jiang, X. Mitochondria-mediated apoptosis in mammals. Protein Cell5, 737–749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sidarala, V. et al. Mitofusin 1 and 2 regulation of mitochondrial DNA content is a critical determinant of glucose homeostasis. Nat. Commun.13, 2340 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anello, M. et al. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia48, 282–289 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Masini, M. et al. Ultrastructural alterations of pancreatic beta cells in human diabetes mellitus.Diabetes Metab. Res. Rev.33, e2894 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Ma, Z. A., Zhao, Z. & Turk, J. Mitochondrial dysfunction and β-cell failure in type 2 diabetes mellitus. Exp. Diabetes Res.2012, 703538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wigger, L. et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat. Metab.3, 1017–1031 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Su, C. et al. 3D chromatin maps of the human pancreas reveal lineage-specific regulatory architecture of T2D risk. Cell Metab.34, 1394–1409 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraja, A. T. et al. Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am. J. Hum. Genet.104, 112–138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonova-Doing, E. et al. An atlas of mitochondrial DNA genotype–phenotype associations in the UK Biobank. Nat. Genet.53, 982–993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer, C. J. et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab.6, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeck, T. et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab.13, 80–91 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Mercader, J. M. et al. Identification of novel type 2 diabetes candidate genes involved in the crosstalk between the mitochondrial and the insulin signaling systems. PLoS Genet.8, e1003046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangwung, P., Petersen, K. F., Shulman, G. I. & Knowles, J. W. Mitochondrial dysfunction, insulin resistance, and potential genetic implications. Endocrinology161, bqaa017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan, L. et al. Mitochondria-associated proteostasis. Annu. Rev. Biophys.49, 41–67 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Bahr, T., Katuri, J., Liang, T. & Bai, Y. Mitochondrial chaperones in human health and disease. Free Radic. Biol. Med.179, 363–374 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiwary, S., Nandwani, A., Khan, R. & Datta, M. GRP75 mediates endoplasmic reticulum–mitochondria coupling during palmitate-induced pancreatic β-cell apoptosis. J. Biol. Chem.297, 101368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibellini, L. et al. The biology of Lonp1: more than a mitochondrial protease. Int. Rev. Cell Mol. Biol.354, 1–61 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Xu, Z. et al. Disuse-associated loss of the protease LONP1 in muscle impairs mitochondrial function and causes reduced skeletal muscle mass and strength. Nat. Commun.13, 894 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, J., Herrmann, J. M. & Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol.22, 54–70 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Quiros, P. M. et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep.8, 542–556 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Shin, C.-S. et al. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat. Commun.12, 265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rep, M. et al. Promotion of mitochondrial membrane complex assembly by a proteolytically inactive yeast Lon. Science274, 103–106 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Cnop, M. et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes63, 1978–1993 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Pollecker, K., Sylvester, M. & Voos, W. Proteomic analysis demonstrates the role of the quality control protease LONP1 in mitochondrial protein aggregation. J. Biol. Chem.297, 101134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uoselis, L. et al. Temporal landscape of mitochondrial proteostasis governed by the UPRmt. Sci. Adv.9, eadh8228 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson, D. W. & McMurray, W. C. Triton solubilization of proteins from pig liver mitochondrial membranes. Biochim. Biophys. Acta856, 515–525 (1986). [DOI] [PubMed] [Google Scholar]
- 35.Slinde, E. & Flatmark, T. Effect of the hydrophile-lipophile balance of non-ionic detergents (Triton X-series) on the solubilization of biological membranes and their integral b-type cytochromes. Biochim. Biophys. Acta455, 796–805 (1976). [DOI] [PubMed] [Google Scholar]
- 36.Smith, L. J., Bolsinger, M. M., Chau, K.-Y., Gegg, M. E. & Schapira, A. H. V. The GBA variant E326K is associated with alpha-synuclein aggregation and lipid droplet accumulation in human cell lines. Hum. Mol. Genet.32, 773–789 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stocki, P., Chapman, D. C., Beach, L. A. & Williams, D. B. Depletion of cyclophilins B and C leads to dysregulation of endoplasmic reticulum redox homeostasis. J. Biol. Chem.289, 23086–23096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochberg, G. K. et al. The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc. Natl Acad. Sci. USA111, E1562–E1570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res.49, D1541–D1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moss, N. D. & Sussel, L. mRNA processing: an emerging frontier in the regulation of pancreatic β cell function. Front. Genet.11, 983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvelos, M. I., Juan-Mateu, J., Colli, M. L., Turatsinze, J.-V. & Eizirik, D. L. When one becomes many—alternative splicing in β-cell function and failure. Diabetes Obes. Metab.20, 77–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bomba-Warczak, E., Edassery, S. L., Hark, T. J. & Savas, J. N. Long-lived mitochondrial cristae proteins in mouse heart and brain. J. Cell Biol.220, e202005193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein, S. H. et al. The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood119, 3321–3329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, C. K., Suda, K., Wang, N. & Schatz, G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science264, 273–276 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Lee, J. et al. Inhibition of mitochondrial LonP1 protease by allosteric blockade of ATP binding and hydrolysis via CDDO and its derivatives. J. Biol. Chem.298, 101719 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peter, B. et al. Defective mitochondrial protease LonP1 can cause classical mitochondrial disease. Hum. Mol. Genet.27, 1743–1753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zurita Rendón, O. & Shoubridge, E. A. LONP1 is required for maturation of a subset of mitochondrial proteins, and its loss elicits an integrated stress response. Mol. Cell. Biol.38, e00412-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandesh, K. et al. Single-cell decoding of human islet cell type-specific alterations in type 2 diabetes reveals converging genetic- and state-driven β-cell gene expression defects. Preprint at bioRxiv10.1101/2025.01.17.633590 (2025).
- 49.Walker, E. M. et al. Retrograde mitochondrial signaling governs the identity and maturity of metabolic tissues.Science388, eadf2034 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christensen, A. A. & Gannon, M. The beta cell in type 2 diabetes. Curr. Diab. Rep.19, 81 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Kim-Muller, J. Y. et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat. Commun.7, 12631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutter, G. A., Sidarala, V., Kaufman, B. A. & Soleimanpour, S. A. Mitochondrial metabolism and dynamics in pancreatic beta cell glucose sensing. Biochem. J.480, 773–789 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Lin, M. T. & Beal, M. F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature443, 787–795 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Zhao, K., Huang, X., Zhao, W., Lu, B. & Yang, Z. LONP1-mediated mitochondrial quality control safeguards metabolic shifts in heart development. Development149, dev200458 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Baumel-Alterzon, S. et al. NRF2 is required for neonatal mouse beta cell growth by maintaining redox balance and promoting mitochondrial biogenesis and function. Diabetologia67, 547–560 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stancill, J. S. & Corbett, J. A. The role of thioredoxin/peroxiredoxin in the β-cell defense against oxidative damage. Front. Endocrinol.12, 718235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gurgul, E., Lortz, S., Tiedge, M., Jörns, A. & Lenzen, S. Mitochondrial catalase overexpression protects insulin-producing cells against toxicity of reactive oxygen species and proinflammatory cytokines. Diabetes53, 2271–2280 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Dai, D.-F. et al. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ. Res.108, 837–846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lang, A. L. et al. A defect in mitochondrial complex III but not in complexes I or IV causes early β-cell dysfunction and hyperglycemia in mice. Diabetes72, 1262–1276 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bai, M. et al. LONP1 targets HMGCS2 to protect mitochondrial function and attenuate chronic kidney disease. EMBO Mol. Med.15, e16581 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kunová, N. et al. The role of Lon-mediated proteolysis in the dynamics of mitochondrial nucleic acid–protein complexes. Sci. Rep.7, 631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wadhwa, R. et al. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res.60, 6818–6821 (2000). [PubMed] [Google Scholar]
- 63.Nanda, S. K., Johnson, R. F., Liu, Q. & Leibowitz, J. L. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′ untranslated region of the Murine hepatitis virus genome. Arch. Virol.149, 93–111 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lytrivi, M., Castell, A.-L., Poitout, V. & Cnop, M. Recent insights into mechanisms of β-cell lipo- and glucolipotoxicity in type 2 diabetes. J. Mol. Biol.432, 1514–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su, M. et al. In silico and in vivo pharmacokinetic evaluation of 84-B10, a novel drug candidate against acute kidney injury and chronic kidney disease. Molecules29, 159 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saunders, D. C. et al. Ectonucleoside triphosphate diphosphohydrolase-3 antibody targets adult human pancreatic β cells for in vitro and in vivo analysis. Cell Metab.29, 745–754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pinti, M. et al. Functional characterization of the promoter of the human Lon protease gene. Mitochondrion11, 200–206 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Bahat, A. et al. Transcriptional activation of LON Gene by a new form of mitochondrial stress: a role for the nuclear respiratory factor 2 in StAR overload response (SOR). Mol. Cell. Endocrinol.408, 62–72 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Fukuda, R. et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell129, 111–122 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Fiorese, C. J. et al. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol.26, 2037–2043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Juliana, C. A. et al. ATF5 regulates β-cell survival during stress. Proc. Natl Acad. Sci. USA114, 1341–1346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolic, J. et al. Proteomic predictors of individualized nutrient-specific insulin secretion in health and disease. Cell Metab.36, 1619–1633 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saunders, D. C. et al. Pancreatlas: applying an adaptable framework to map the human pancreas in health and disease. Patterns1, 100120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, Y. J. et al. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metab.24, 616–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Georgiadou, E. & Rutter, G. A. Control by Ca2+ of mitochondrial structure and function in pancreatic β-cells. Cell Calcium91, 102282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balderas, E. et al. Mitochondrial calcium uniporter stabilization preserves energetic homeostasis during complex I impairment. Nat. Commun.13, 2769 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tangeda, V. et al. Lon upregulation contributes to cisplatin resistance by triggering NCLX-mediated mitochondrial Ca2+ release in cancer cells. Cell Death Dis.13, 241 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pearson, G. L., Gingerich, M. A., Walker, E. M., Biden, T. J. & Soleimanpour, S. A. A selective look at autophagy in pancreatic β-cells. Diabetes70, 1229–1241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ozkurede, U. et al. Cap-independent mRNA translation is upregulated in long-lived endocrine mutant mice. J. Mol. Endocrinol.63, 123–138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoshino, A. et al. Oxidative post-translational modifications develop LONP1 dysfunction in pressure overload heart failure. Circ. Heart Fail.7, 500–509 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Wu, L. et al. Sirt3 restricts tumor initiation via promoting LONP1 deacetylation and K63 ubiquitination. J. Transl. Med.21, 81 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shin, M. et al. Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Nat. Commun.12, 3239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cadavez, L. et al. Chaperones ameliorate beta cell dysfunction associated with human islet amyloid polypeptide overexpression. PLoS ONE9, e101797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Engin, F. & Hotamisligil, G. S. Restoring endoplasmic reticulum function by chemical chaperones: an emerging therapeutic approach for metabolic diseases. Diabetes Obes. Metab.12, 108–115 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Bar-Ziv, R. et al. Glial-derived mitochondrial signals affect neuronal proteostasis and aging. Sci. Adv.9, eadi1411 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodríguez, C. I. et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet.25, 139–140 (2000). [DOI] [PubMed] [Google Scholar]
- 87.Sidarala, V. et al. Mitophagy protects β cells from inflammatory damage in diabetes. JCI Insight5, e141138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Corsa, C. A. S. et al. The E3 ubiquitin ligase parkin is dispensable for metabolic homeostasis in murine pancreatic β cells and adipocytes. J. Biol. Chem.294, 7296–7307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thorens, B. et al. Ins1Cre knock-in mice for beta cell-specific gene recombination. Diabetologia58, 558–565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pearson, G. et al. Clec16a, Nrdp1, and USP8 form a ubiquitin-dependent tripartite complex that regulates β-cell mitophagy. Diabetes67, 265–277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kamat, V. et al. A versatile pumpless multi-channel fluidics system for maintenance and real-time functional assessment of tissue and cells. Cell Rep. Methods3, 100642 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wittig, I., Braun, H. P. & Schagger, H. Blue native PAGE. Nat. Protoc.1, 418–428 (2006). [DOI] [PubMed] [Google Scholar]
- 93.Chaudhry, A., Shi, R. & Luciani, D. S. A pipeline for multidimensional confocal analysis of mitochondrial morphology, function, and dynamics in pancreatic β-cells. Am. J. Physiol. Endocrinol. Metab.318, E87–E101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soleimanpour, S. A. et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell157, 1577–1590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paulo, J. A. Isobaric labeling: expanding the breadth, accuracy, depth, and diversity of sample multiplexing. Proteomics22, e2200328 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diraison, F. et al. Over-expression of sterol-regulatory-element-binding protein-1c (SREBP1c) in rat pancreatic islets induces lipogenesis and decreases glucose-stimulated insulin release: modulation by 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR). Biochem. J.378, 769–778 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gingerich, M. A. et al. An intrinsically disordered protein region encoded by the human disease gene CLEC16A regulates mitophagy. Autophagy19, 525–543 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ondrovicová, G. et al. Cleavage site selection within a folded substrate by the ATP-dependent lon protease. J. Biol. Chem.280, 25103–25110 (2005). [DOI] [PubMed] [Google Scholar]
- 99.Motulsky, H. J. & Brown, R. E. Detecting outliers when fitting data with nonlinear regression—a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics7, 123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–4 and Fig. 1.
Protein identifiers and quantitative measures in the insoluble fractions of human islets.
Protein identifiers and quantitative measures in the soluble fractions of human islets.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Statistical source data.
Statistical source data and unprocessed western blots.
Statistical source data.
Statistical source data.
Statistical source data and unprocessed immunoblots.
Statistical source data.
Statistical source data.
Statistical source data and unprocessed immunoblots.
Statistical source data and unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Unprocessed immunoblots.
Data Availability Statement
The proteomics data have been deposited and are publicly available at the ProteomeXchange (PXD060404). Microscopy data will be shared by the lead contact upon reasonable request. Any additional information required to reanalyse the data reported in this paper is available from the lead author upon reasonable request. Source data are provided with this paper.