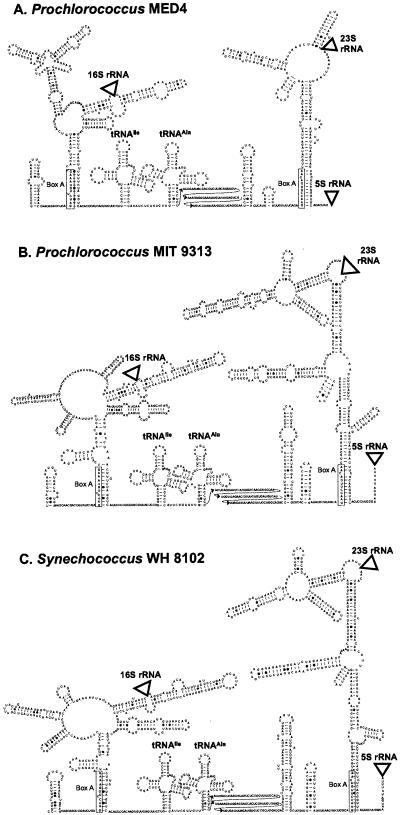
FIG. 2.
Predicted secondary structures of the ITS regions of the rRNA operon in (A) Prochlorococcus strain MED4, (B) Prochlorococcus strain MIT 9313, and (C) Synechococcus strain WH 8102. Locations of the 16S rRNA, 23S rRNA, and 5S rRNA are represented by triangles. All strains have antiterminator box B loops and box A sequences conserved in most bacterial ITS sequences, as well as genes for tRNAs isoleucine and alanine typical of cyanobacterial ITS sequences. Sequence corresponding to the box A motif is enclosed in a rectangle. The 5′ region of the tRNAAla-23S rRNA spacer (between the tRNAAla and the box B loop) for which no structure was inferred is shown in three rows of text to save space.