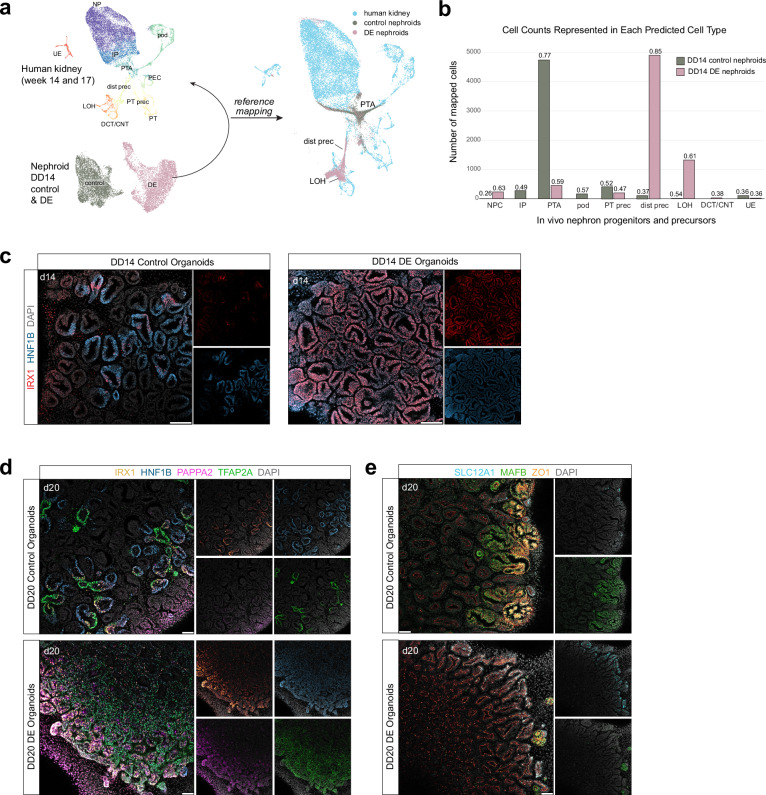
Fig. 3. Characterizations of distal nephron segmentation in distal-enriched organoids.
a Schematic showing reference mapping of DD14 organoid scRNA sequencing dataset to reanalyzed week 14 and week 17 (4772 cells) merged human nephron and ureteric scRNA sequencing data1,49. (Right) UMAP of human kidney (reference) overlaid with DD14 control and DE organoid mapping. b Bar plot showing number of organoid cells mapping to each predicted cell type, split by condition. Numbers above bars denote prediction score for each cell type. c Immunofluorescence stain of DD14 control and DE organoids showing detection of IRX1. Biological replicates (n = 3) showed similar results. Scale bar: 50 microns. d–e Immunofluorescent stains of DD20 control and DE organoids showing detection of select loop of Henle/macula densa precursor and podocyte precursor markers. Biological replicates (n = 3) showed similar results. Scale bar: 50 microns. Abbreviations: NP nephron progenitors, IP induced progenitors, PTA pretubular aggregate, pod podocytes, PEC parietal epithelial cells, dist prec distal nephron precursors, PT prec proximal tubule precursors, PT proximal tubule, LOH loop of Henle, DCT/CNT distal convoluted tubule/connecting tubule.