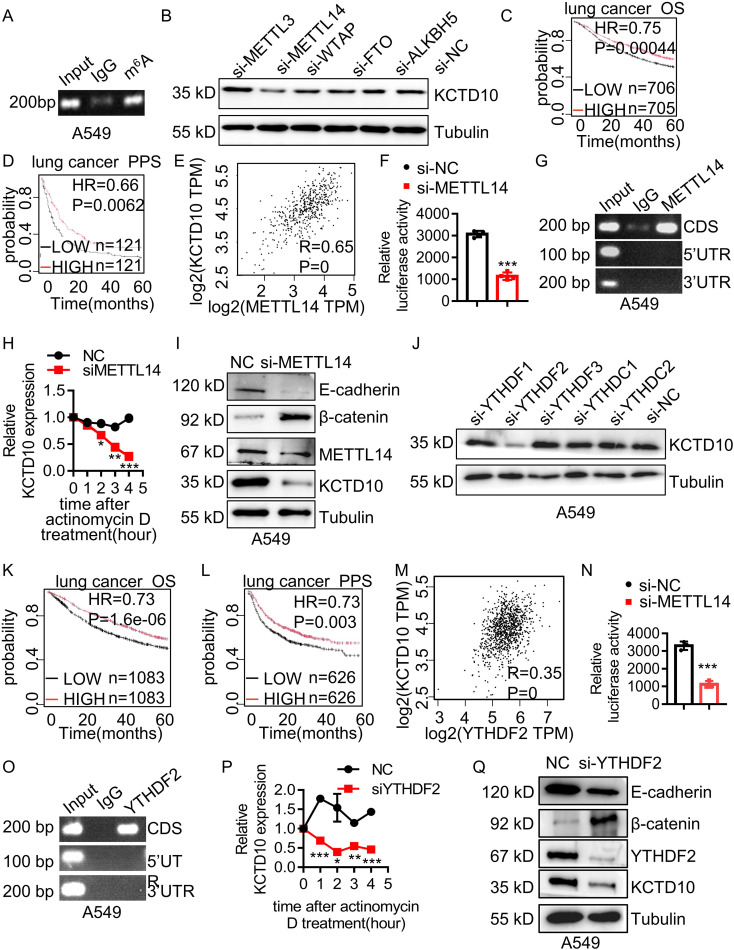
Figure 8.
METTL14 and YTHDF2 mediates m6A modification of KCTD10 and enhances its mRNA stability. (A) MeRIP detecting m6A modification of KCTD10 CDS. (B) KCTD10 protein levels following m6A-related interfering RNAs. (C, D) Correlation between METTL14 expression and overall/post-progression survival in lung cancer patients using Kaplan-Meier Plotter survival analysis. (E) Correlation between METTL14 and KCTD10 expression in lung cancer using the GEPIA database. (F) Effects of METTL14 on the luciferase reporter activity of KCTD10. (G) RIP analysis detecting METTL14 binding to the predicted modification site of KCTD10. (H) qPCR analysis of KCTD10 RNA stability following METTL14 knockdown. (I) Western blot analysis of KCTD10 and related protein expression in A549 cells following METTL14 interference. (J) KCTD10 protein expression after m6A-related RNA interference. (K, L) Correlation between YTHDF2 expression and patient survival. (M) The GEPIA database analyzing the correlation between YTHDF2 and KCTD10 expression. (N) Effect of YTHDF2 knockdown on luciferase reporter activity of KCTD10. (O) RIP analysis of YTHDF2 binding to predicted modification sites of KCTD10 in A549 cells. (P) qPCR analysis of KCTD10 RNA stability following YTHDF2 knockdown. (Q) Western blot analysis of KCTD10 and downstream gene expression following YTHDF2 knockdown. *P < 0.05, **P < 0.01, ***P < 0.001.