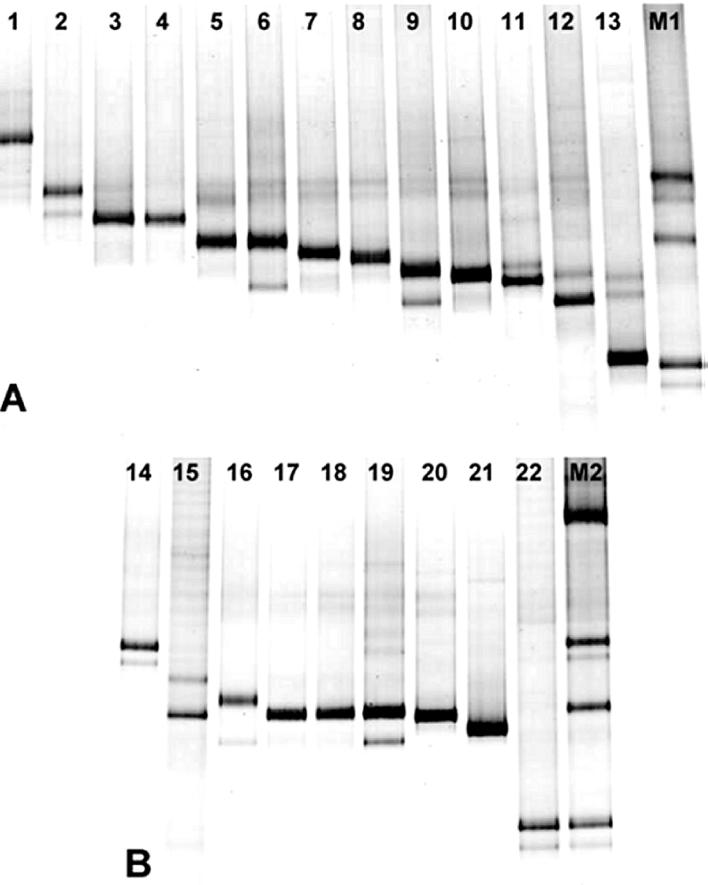
FIG. 2.
DGGE patterns of 16S rDNA fragments of Burkholderia species (A) and members of the B. cepacia complex (B) generated by PCR with Burkholderia-specific primers (positions 155 to 663 [E. coli numbering] [10]) in a 50 to 60% denaturing gradient. Lane 1, B. glathei WD1; lanes 2 and 14, B. multivorans LMG13010; lane 3, B. plantarii NCPPB3590; lane 4, B. gladioli ATCC 33664; lane 5, B. pyrrocinia ATCC 15958; lanes 6 and 16, B. stabilis LMG14294; lanes 7 and 18, B. vietnamiensis LMG10929; lane 8, B. phenazinium LMG2247; lane 9, B. caribensis WD3; lane 10, B. glumae NCPPB3708; lane 11, B. graminis WD2; lane 12, B. caribensis LMG18531; lane 13, B. caryophylli NCPPB353; lane 15, B. cepacia LMG16656 (genomovar III); lane 17, B. cepacia ATCC 25416 (genomovar I); lane 19, B. cepacia NCPPB945; lane 20, B. cepacia P2; lane 21, B. cepacia IPO1718; lane 22, B. cepacia LMG18941 (genomovar VI); lane M1, Burkholderia marker containing (from top to bottom) B. multivorans LMG13010, B. cepacia ATCC 25416, and B. cepacia LMG18941; lane M2, Burkholderia marker containing (from top to bottom) B. andropogonis LMG6872, B. multivorans LMG13010, B. cepacia ATCC 25416, and B. cepacia LMG18941.