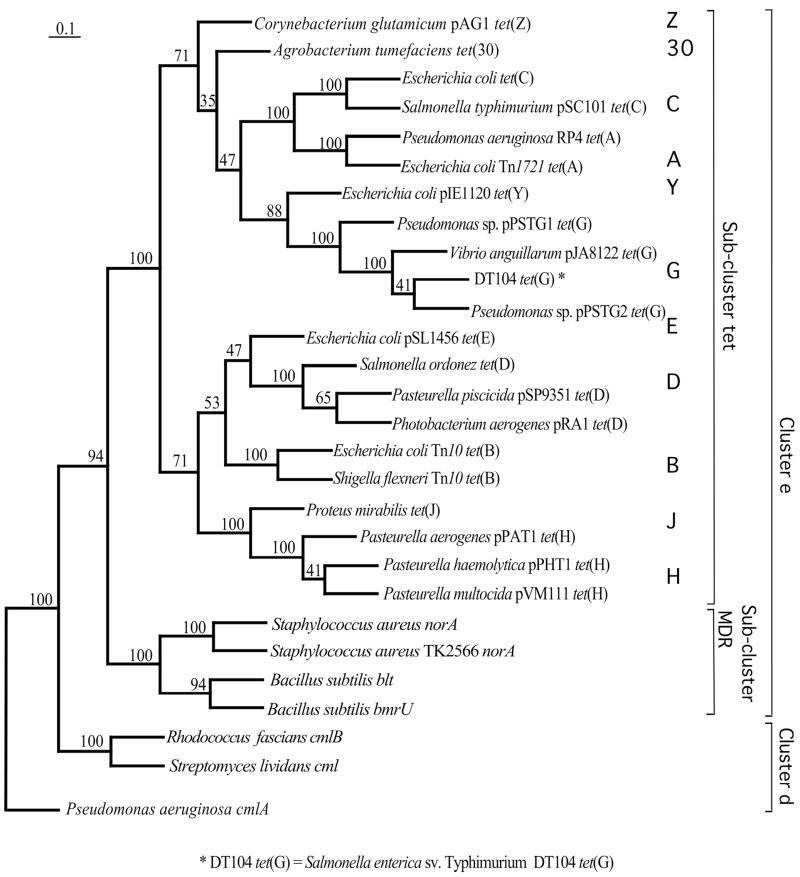
FIG. 1.
Maximum-likelihood tree of tetracycline resistance genes, encoding tet efflux pumps of gram-negative bacteria (subcluster tet), and other genes of the DHA12 family. The same overall topology was obtained by neighbor-joining and parsimony analyses. Numbers at nodes represent the percentages of occurrence of nodes in 100 bootstrap trials. The scale bar is in fixed nucleotide substitutions per sequence position. The tree is arbitrarily rooted with the P. aeruginosa cmlA gene. The set of PCR primers, presented in Table 2, targets various classes of tet genes, which are shown in boldface.