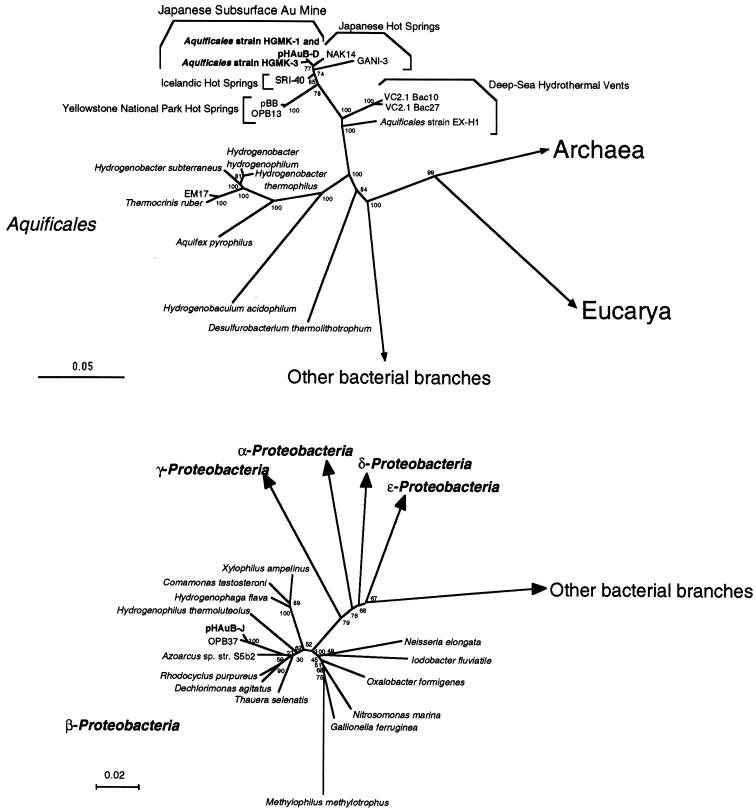
FIG. 1.
Phylogenetic trees of representative Aquificales strains and phylotypes (upper panel) and of beta subdivision Proteobacteria strains and phylotypes (lower panel). Each of the trees was inferred by neighbor-joining analysis of approximately 600 homologous positions of the rDNA sequence. Environmental clones VC2.1 Bac10 and VC2.1 Bac27 were obtained from deep-sea hydrothermal vent environments (25, 27), and EM17, pBB, OPB13, and OPB37 were from Yellowstone National Park hot spring communities (12, 26, 28). The clones SRI-40, NAK14, and GANI-3 were recovered from Icelandic and Japanese hot spring microbial mats (29, 32, 40). Bootstrap analysis was performed with 100 resampled data sets. Bar, 0.05 (upper panel) or 0.02 (lower panel) change per nucleotide. The accession numbers of the 16S rDNA sequences in this figure are: Desulfurobacterium thermolithotrophicum, AJ001049; Hydrogenobaculum acidophilus, D16296; Aquifex pyrophilus, M83548; Thermocrinis ruber, AJ005640; EM17, U05661; Hydrogenobacter subterraneus, AB026268; Hydrogenobacter thermophilus, Z30214; Hydrogenobacter hydrogenophilum, Z30242; Aquificales sp. strain EX-H1, AF188332; VC2.1 Bac10, AF068791; VC2.1 Bac27, AF068801; pBB, AF113542; OPB13, AF027098; SRI-40, AF255598; NAK14, AB005738; GANI-3, AB005735; Aquificales sp. strain HGM-K1, AB071324; strain HGM-K3, AB071325; pHAuB-D, AB071326; pHAuB-J, AB071327; OPB37, AF026985; Xylophilus ampelinus, AF078758; Comamonas testosteroni, M11224; Hydrogenophaga flava, AF078771; Hydrogenophilus thermoluteolus, AB009828; Azoarcus sp. strain S5b2, L15532; Rhodocyclus purpureus, M34132; Dechlorimonas agitatus, AF047462; Thauera selenatis, X68491; Methylophilus methylotrophus, M29021; Gallionella ferruginea, L07897; Nitrosomonas marina, Z46990; Oxalobacter fornigenes, U49757; Iodobacter fluviatile, M22511; and Neisseria elongata, L06171.