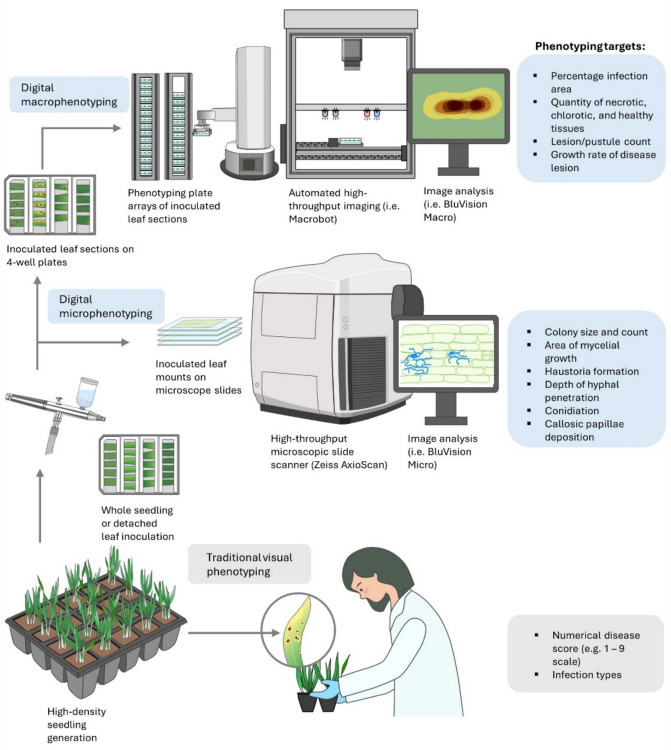
Fig. 1.
Comparison of traditional visual phenotyping of disease resistance to image-based macro- and micro-phenotyping. Phenotyping using the human eye is limited to numerical scores and infection types, whereas digital phenotyping can quantitatively measure multiple aspects of the host–pathogen interaction resistance macro- and microscopically. Image-based phenotyping workflows, such as those used at the IPK (Germany) and most recently deployed at La Trobe University (Australia), involve an initial controlled inoculation either on detached leaves or as whole seedlings that are mounted onto four wells (containing water agar, silver nitrate, and a senescence inhibitor) to image live leaves during the progression of disease development. A robotic arm automatically loads plates from magazines with a capacity sufficient to hold a whole experiment. Images are acquired with a monochrome digital camera on a stage selectively illuminated by LED lights with peak wavelengths 365 nm (UV), 470 nm (blue), 530 nm (green), and 625 nm (red). The standardized images are analyzed by dedicated software modules published on open-source GitHub repositories that can be modified for specific pathogens (Lück et al. 2020). Digital microphenotyping workflows such as “BluVision Micro” rely upon commercially available slide scanners common in medical pathology to reduce laborious sample handling and acquisition such as the Zeiss® Axioscan (Carl Zeiss Group). These microscopic workflows rely upon specific staining steps to facilitate ease of visualization and subsequent image analysis, and the choice of stain is highly dependent upon the pathosystem under investigation