Abstract
Mitochondrial DNA (mtDNA) base editors are powerful tools for investigating mitochondrial diseases. However, their editing efficiency can vary significantly depending on the target site within the mtDNA. In this study, we developed two improved versions of the mitochondrial adenine base editor (Hifi-sTALED and αnHifi-sTALED) by modifying components other than the TadA8e-V28R deaminase variant. These enhancements significantly increased editing efficiency while preserving minimal off-target effects across the transcriptome. Using these optimized editors, we achieved improved mtDNA editing in mouse embryos and successfully generated mt-Rnr1 mutant mice with high heteroplasmic loads. Functional analyses revealed that the mt-Rnr1 mutation impaired mitochondrial function, as indicated by reduced ATP production and decreased oxygen consumption rate (OCR). These findings demonstrate the utility of the enhanced base editors in generating mitochondrial disease models and advancing research in mitochondrial genetics.
Keywords: MT: RNA/DNA Editing, mitochondria, mtDNA, mitochondrial editing, TALED, base editing
Graphical abstract

Lee and colleagues developed two enhanced mitochondrial adenine base editors, Hifi-sTALED and αnHifi-sTALED, that improve editing efficiency while maintaining minimized off-target effects. These tools successfully generated mt-Rnr1 mutant mice and revealed mitochondrial dysfunction, offering a robust platform for modeling mitochondrial diseases and studying mtDNA mutations.
Introduction
Mitochondria play a central role in cellular energy production and carry out a range of essential biological functions, supported by their own genome, which encodes key tRNAs, rRNAs, and components of the respiratory complexes.1 Mutations in mitochondrial DNA (mtDNA) can lead to significant functional impairments and are associated with a wide range of severe mitochondrial diseases.2 The development of TALE-guided mitochondrial base editors3,4 has enabled the generation of genetically modified models, such as mice carrying mtDNA mutations, thereby advancing the study of mitochondrial disorders.5,6 Previous work on mitochondrial adenine base editors, such as TALED (TALE-linked deaminase), demonstrated their potential to introduce mtDNA mutations in mouse models while minimizing transcriptome-wide off-target effects through the use of the TadA8e-V28R deaminase variant.6 However, limited editing efficiency, especially in regions with low target accessibility, has constrained broader applications, including the creation of diverse disease models.
Building on recent findings, our study sought to optimize the non-deaminase components of TALED to enhance editing activity while preserving high specificity. This effort led to the development of two improved editors, Hifi-sTALED and αnHifi-sTALED, designed to improve editing efficiency and enable the efficient generation of disease-relevant mtDNA mutations. Here, we report the performance of these enhanced tools in producing mtDNA mutations in mouse models, providing a foundation for more effective investigations into mitochondrial dysfunction and disease.
Results
Alterations in sTALED enhance editing efficiency
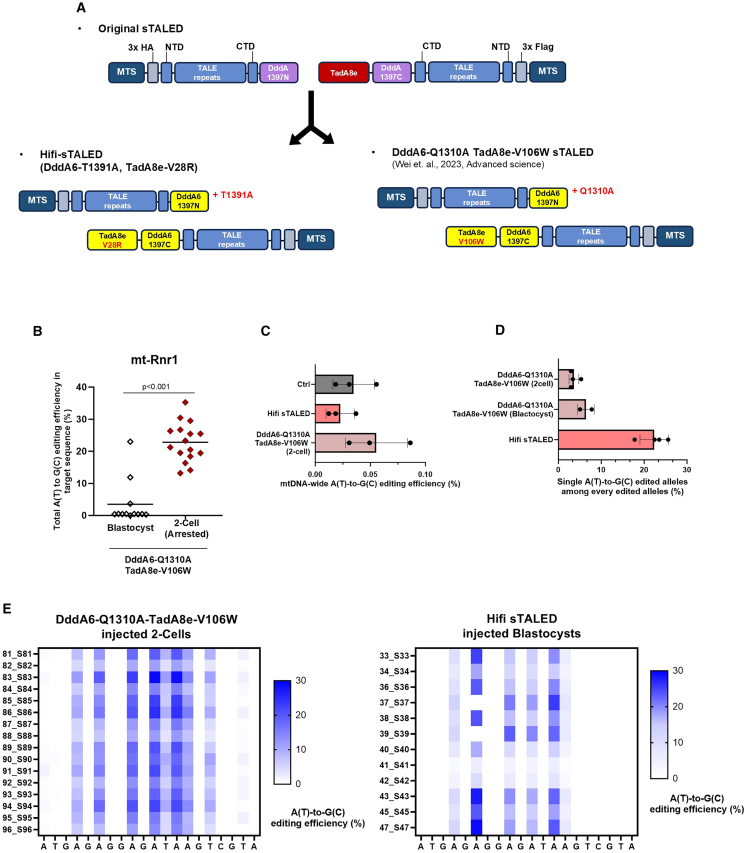
sTALED consists of four key components: a mitochondrial translocation sequence (MTS) for mitochondrial targeting, TALE repeats for DNA recognition, a DddA (half) domain for double-stranded DNA unwinding, and TadA8e for adenine deamination. Because TadA8e requires single-stranded DNA for deamination, the DNA unwinding activity of the DddA (half) domain directly influences the editing efficiency of sTALED. Among various DddA (half) variants, DddA6 has been reported to exhibit the highest efficiency7 and has shown optimal performance when combined with sTALED.8
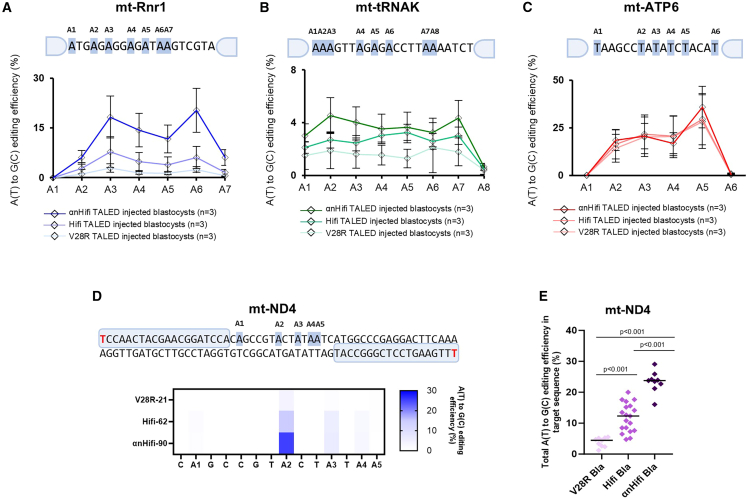
However, increased activity of the DddA6 variant in TALED may lead to higher off-target editing in mtDNA.9 To mitigate this, we introduced the T1391A mutation, known to significantly reduce off-target DNA editing,9 resulting in a modified editor termed “Hifi-sTALED” (Figure 1A). Additionally, it has been shown that altering the N-terminal region of TALE repeats (alpha N-terminal) in DdCBE (a mitochondrial cytosine base editor) can change the 5′ base-recognition pattern.10 Based on this, we hypothesized that introducing this alpha N-terminal configuration could enhance the overall DNA-binding affinity of sTALED and improve its editing efficiency. This second variant is referred to as “αnHifi-sTALED.” Both of these enhanced editors were built upon the TadA8e-V28R variant, which is known to reduce transcriptome-wide off-target effects, with the goal of generating mtDNA mutant mice. These editors were then tested on three mtDNA target sites in mice. Two of the targets (mt-Rnr1 and mt-tRNAK) showed low editing efficiency with V28R-sTALED, making it unlikely that disease-related phenotypes would be observed in the resulting mouse models. The third target, mt-ATP6, which had already demonstrated high editing efficiency with V28R-sTALED (mt-ATP6),6 was included for comparison (Figure 1B).
Figure 1.
Modifying sTALED to enhance editing efficiency
(A) Concept of progress in our study. The components are also described in the results and discussion sections. DddA6h refers to DddA6 variants with the T1391A (Hifi) variant. (B) Target sequences in mouse mitochondrial DNA for A(T)-to-G(C) conversion. (C–E) Total A(T)-to-G(C) editing efficiency in the target sequences of blastocysts using three versions of sTALED. Average efficiency is indicated by bars. p values were calculated via Student’s two-tailed t test.
To evaluate editing performance, we microinjected mRNA encoding each of the three sTALED types (V28R-, Hifi-, and αnHifi-sTALED) into mouse zygotes and performed targeted deep sequencing after incubation to the blastocyst stage (Figures 1C–1E). Both Hifi- and αnHifi-sTALED exhibited higher average A(T)-to-G(C) editing rates across all three target sites compared to V28R-sTALED. At the mt-Rnr1 site, αnHifi-sTALED achieved a 5.4-fold increase in editing efficiency over V28R-sTALED (Figure 1D) and a 2.1-fold increase over Hifi-sTALED. In contrast, at the mt-ATP6 target, no significant improvements were observed, with only marginal increases in editing efficiency (44.2%, 44.7%, and 48.3% for V28R-, Hifi-, and αnHifi-sTALED, respectively). Since mitochondrial base editing efficiency is closely linked to mtDNA replication,11 embryonic injection of mRNA may have resulted in a saturation point, limiting further increases in editing efficiency (Figure 1E).
Improved editing efficiency through V28R-based modifications does not alter the editing pattern
Although modifying sTALED components enhanced editing efficiency, it did not change the overall editing pattern within the target window (Figures 2A–2C). Because the TadA8e-V28R deaminase exhibits a single-base editing preference,6 allele analysis of edited blastocysts confirmed that both enhanced versions of sTALED maintained this same specificity (Figure S1).
Figure 2.
Analysis of editing patterns with Hifi-sTALED and αnHifi-sTALED
(A–C) A(T)-to-G(C) edited sites in target sequences with the edited percentages of blastocysts. Since no notable editing occurred in the opposite strand, only adenines in edited strands are displayed. Error bars represent standard deviation (SD). (D) Target sequence in mouse mitochondrial mt-ND4 and editing patterns of mt-ND4 edited blastocysts. (E) Total A(T)-to-G(C) editing efficiency in the mt-ND4 target of blastocysts. Average efficiency is indicated by bar. p values were calculated via Student’s two-tailed t test.
We next investigated whether the enhanced tools could improve editing efficiency at specific nucleotide positions within the editing window, sites characterized by position-specific base editing rather than broad editing activity. The mouse mt-ND4 target site is one such example, displaying low overall editing efficiency with V28R-sTALED but a consistent editing preference at the A2 position. To evaluate this, we injected embryos with mt-ND4-targeting Hifi- and αnHifi-sTALED mRNA and analyzed editing outcomes in the resulting blastocysts (Figure 2D; Figure S2). The A(T)-to-G(C) editing efficiencies at the A2 site and across the entire target region were 3.7%/5.5% for V28R-sTALED, 15.0%/20.0% for Hifi-sTALED, and 25.3%/29.9% for αnHifi-sTALED. These results demonstrate that the enhanced editors not only improve overall editing across the target site (Figure 2E) but also substantially increase editing efficiency at specific nucleotide positions. Moreover, although the TALE alpha N-terminal is known to improve TALE binding at sequences that do not begin with a 5′T, αnHifi-sTALED still exhibited the highest editing efficiency at the mt-ND4 site, where both the left and right TALE-binding sequences begin with 5′T. This suggests that αnHifi-sTALED offers improved performance even at traditionally favorable TALE-binding contexts.
DddA6-T1391A and TadA8e-V28R variants mediate relatively precise targeting
A previous study reported that extensive modifications to sTALED, specifically incorporating the Q1310A mutation in DddA6 and the V106W mutation in TadA8e, substantially improved editing efficiency and proposed this combination as an optimized version of sTALED.8 To compare this previously reported variant (DddA6-Q1310A/V106W-TadA8e-sTALED) with our version (Hifi-sTALED: T1391A-DddA6 and V28R-TadA8e), we targeted the mt-Rnr1 locus using each editor and performed embryo injections followed by analysis (Figure 3A).
Figure 3.
Comparison of editing activity between Hifi-sTALED and DddA6-Q1310A–TadA8e-V106W-sTALED
(A) Schematic of Hifi-sTALED and DddA6-Q1310A–TadA8e-V106W sTALED. (B) Total A(T)-to-G(C) editing efficiency in the mt-ND4 target of blastocysts. Average efficiency is indicated by a bar. p values were calculated via Student’s two-tailed t test. (C) Whole mtDNA A(T)-to-G(C) conversion rates of untreated blastocyst, DddA-T1391A TadA8e-V28R sTALED-treated blastocyst, and DddA6-Q1310A-TadA8e-V106W sTALED-treated two-cell stage arrested embryos. Only conversions with frequencies >1% are included, excluding those in the target sequence. (D) Frequencies of single A(T)-to-G(C) edited alleles relative to total edited alleles. Error bars represent SD. (E) Editing patterns in the mt-Rnr1 target of embryos injected with DddA6-Q1310A–TadA8e-V106W sTALED (two-cell stage arrested) and Hifi-sTALED (blastocyst stage). Each item on the y axis of the heatmap represents a single embryo (two-cell or blastocyst stage).
Notably, several embryos injected with DddA6-Q1310A/V106W-TadA8e-sTALED failed to progress beyond the 2-cell stage, even after incubation to the blastocyst stage (Figure 3B). Among the embryos that developed to the blastocyst stage, only 3 out of 12 exhibited editing, whereas all 16 of the arrested two-cell embryos exhibited targeted editing. Given our previous findings that the TadA8e-V106W variant induces transcriptome-wide off-target effects at levels similar to unmodified TadA8e,6 this developmental arrest may be attributed to these off-target effects.
However, a genome-wide analysis of mtDNA off-target editing revealed no significant differences among the variants (Figure 3C). This is likely because sTALED-mediated editing is predominantly driven by TadA8e activity, and the intrinsic adenine deamination activity of V106W and V28R variants does not differ substantially.
We previously examined the editing profiles of V28R-TadA8e at the target site and found that it generated a higher proportion of single-base edited alleles compared to unmodified TadA8e.6 To directly compare this with the V106W variant, we calculated the ratio of single-base edited alleles to the total number of edited alleles. This ratio was significantly higher for Hifi-sTALED (22.33%) than for DddA6-Q1310A/V106W-TadA8e-sTALED (3.62%) (Figure 3D). Additionally, DddA6-Q1310A/V106W-TadA8e-sTALED edited adenines on both DNA strands within the target region, whereas Hifi-sTALED showed strand-specific activity (Figure 3E). These results suggest that Hifi-sTALED mediates more precise and strand-selective base editing.
Assessing mutant mouse generation with Hifi- and αnHifi-sTALED
Although embryos injected with TALED variants developed normally to the blastocyst stage, increased base editing activity could potentially lead to embryonic lethality. To determine whether our enhanced sTALED variants could generate mutant mice without inducing developmental toxicity, we performed embryo transfer using the injected embryos.
mt-Rnr1 mutant mice were successfully generated using both Hifi- and αnHifi-sTALED. Notably, αnHifi-sTALED achieved a target site editing efficiency of up to 62.87% in mt-Rnr1 mutant mice (Figure 4A), and germline transmission of the mt-Rnr1 mutation was confirmed by genotyping F1 offspring (Figure 4B).
Figure 4.
Generation of mitochondrial mutant mouse models using modified sTALEDs
(A) Total A(T)-to-G(C) editing efficiency in the mt-Rnr1 target of mutant mice. The dotted bar indicates the average efficiency. (B) Editing patterns in mt-Rnr1 target of mutant-induced mice. (C and D) Whole mtDNA sequencing results of edited mice. (C) Dots represent base conversions with frequencies >1%. Black dots indicate naturally occurring single-nucleotide variations (SNVs), and blue dots indicate conversions in target sequences. SNVs consistently found in all wild-type littermate mice were excluded. (D) Whole mtDNA A(T)-to-G(C) conversion rates of littermate wild-type mice and αNHifi edited mt-Rnr1 mutant mice. Only conversions with frequencies >1% are included, excluding those in the target sequence. p values were calculated via Student’s two-tailed t test.
In a previous study, V28R-sTALED-injected mice exhibited negligible mtDNA-wide off-target effects compared to untreated controls.6 To evaluate whether the enhanced editing activity of αnHifi-sTALED introduced any mtDNA-wide off-target effects, we compared αnHifi-sTALED-injected mice with their unedited littermates. No significant base conversions were observed across the mitochondrial genome (Figure 4C), and no notable A(T)-to-G(C) off-target editing events were detected (Figure 4D).
Phenotypic analysis of mt-Rnr1 mutation
To evaluate the cellular phenotype resulting from the mt-Rnr1 mutation, we generated mt-Rnr1 mutant NIH3T3 cell lines using αnHifi-sTALED targeted to the mt-Rnr1 locus. Briefly, NIH3T3 cells were transfected with αnHifi-sTALED, and single-cell clones were isolated. After incubation for over 30 days, targeted deep sequencing confirmed the stable incorporation of mtDNA mutations (Figure 5A).
Figure 5.
Phenotypes of cell lines and mice harboring mt-Rnr1 mutations
(A) Schematic design for isolating single cells with induced mutations. (B) Total A(T)-to-G(C) editing efficiency in the mt-Rnr1 target of mutant-induced isolated NIH3T3 cells. Dotted bar represents the average efficiency. (C) Oxygen consumption rates (OCRs) in wild-type NIH3T3 cell and mt-Rnr1 mutant cell lines, Error bars represent SD. (D) Extracellular acidification rates (ECARs) in wild-type NIH3T3 cell and mt-Rnr1 mutant cell lines. Error bars represent SD. (E) ATP assay results of mt-Rnr1 mutant cell lines. Gray bar represents wild-type NIH3T3, and the red bar represents mt-Rnr1-edited cell lines (as shown in the figure). (F) Schematic illustration of the overall workflow of the mouse experiment. (G) ATP assay results of mt-Rnr1 mutant mice. (H–J) Western blot results. (H) Protein expression levels of mitochondrial-encoded genes in wild-type and mutant mouse quadriceps tissues were analyzed by western blotting. TOMM20 was used as a loading control. (I) Each band intensity was normalized to TOMM20, and relative quantification was performed by setting the average value of the wild-type group to 1. n = 3 tested. (J) Values from three wild-type mice were grouped as the WT group, and statistical significance was determined using Student’s t test. Signal intensities were measured using ImageJ.
Various heteroplasmic levels were observed among the resulting cell lines, with editing efficiencies reaching up to 96.26% for A(T)-to-G(C) conversions within the target sequence (Figure 5B; Figure S3A). Although editing efficiencies varied among clones, the same nucleotide positions were consistently edited (Figure S3B).
Next, we evaluated the impact of the mt-Rnr1 mutation on mitochondrial function. The TALED-induced editing introduced multiple substitutions in the mt-Rnr1 gene (m.971A>G, m.973A>G, m.976A>G, m.978A>G, m.980A>G, and m.981A>G), a region known to be functionally constrained.12 Notably, the m.978A>G mutation corresponds to the human m.1555A>G variant, which is associated with mitochondrial disease. To investigate the effects of these mutations, we performed oxygen consumption rate (OCR) assays on two mutant cell lines (#6 and #30) with editing efficiencies of 95% and 87%, respectively. OCR was markedly reduced in both mutant lines, with levels nearly undetectable compared to wild-type cells (Figure 5C).
In contrast, extracellular acidification rate (ECAR) was significantly elevated in the mutant cell lines, suggesting a shift toward glycolysis as an alternative metabolic pathway (Figure 5D). Although ATP levels were also reduced, the decline was less pronounced than in OCR measurements. Line #6 showed a statistically significant decrease in ATP (p < 0.002), while line #30 exhibited a decreasing trend that did not reach statistical significance (p = 0.073) (Figure 5E).
We further extended our phenotypic analysis to mt-Rnr1 mutant mice. Given the sensitivity of muscle tissue to mitochondrial dysfunction, quadriceps muscles from 8-week-old male mice were analyzed for ATP content (Figure 5F). The mutant mouse with the highest heteroplasmic load (#005) exhibited the lowest ATP level, while those with lower heteroplasmic rates (#009 and #012) showed ATP levels comparable to unedited controls (heteroplasmy rates of 47.21%, 16.10%, and 1.4%, respectively) (Figure 5G).
Since the introduced mutation targets mt-Rnr1, a component of the mitochondrial ribosome, we performed western blotting to assess mitochondrial protein expression levels in quadriceps muscles. After mitochondrial isolation, western blot analyses were conducted to characterize mitochondrially encoded subunits from several respiratory complexes: ND5 (complex I), CYTB (complex III), COX1 (complex IV), and ATP6 (complex V). Proteins of complex II were excluded, as all its subunits are nuclear-encoded (Figure 5H).
Given that the mt-Rnr1 mutant mice exhibited heteroplasmy rates of 47.21%, 16.10%, and 1.4% for #005, #009, and #012, respectively, and that ATP levels showed an inverse correlation with these rates (Figure 5G), we hypothesized that expression levels of mitochondrially encoded proteins might also inversely correlate with heteroplasmy. However, quantitative analysis of the western blot data revealed no such correlation (Figure 5I). Although no clear overall correlation was observed, comparisons between individual mutant mice and the wild-type group indicated that mutant mice, particularly regarding MT-ND5 and MT-ATP6, exhibited lower protein expression relative to the wild-type average (Figure 5J).
This discrepancy may arise from several factors. For example, compensatory mechanisms at the transcriptional or translational level responding to mitochondrial defects may mask differences at the protein level.13,14 Additionally, cellular remodeling, including transcriptomic changes triggered by mitochondrial dysfunction, is known to vary among individuals,15 which may explain the variability in protein expression observed here.
Discussion
In this study, we successfully developed two enhanced mitochondrial adenine base editors, Hifi-sTALED and αnHifi-sTALED, by modifying components other than the TadA8e deaminase. These modifications were designed to improve mtDNA editing efficiency while preserving the editing profile of TadA8e-V28R. Both variants demonstrated significantly enhanced editing efficiency in mouse blastocysts, facilitating the generation of mt-Rnr1 mutant mice with higher heteroplasmy rates.
Functional analyses revealed that mt-Rnr1 mutations led to mitochondrial dysfunction, as evidenced by reduced OCR and decreased ATP levels. These results establish a link between mt-Rnr1 mutations and mitochondrial defects, thereby contributing to a better understanding of the molecular basis of mitochondrial diseases.
While our study primarily focused on enhancing editing efficiency, recent progress in the development of mtDNA adenine base editors, such as the incorporation of engineered variants like TadA8e-RW and DddA6, has demonstrated further improvements in mtDNA editing in rat models. Moreover, coupling DddA6 with enzymes involved in base excision repair, such as human uracil-DNA glycosylase (hUNG), has been shown to enhance editing efficiency even further.16,17 These findings suggest that integrating αnHifi-sTALED with such emerging variants could yield even higher A(T)-to-G(C) conversion rates.
Despite these advancements, some limitations remain. First, similar to other mitochondrial base editing tools, our enhanced editors do not address the issue of bystander editing. This limitation highlights the need for future studies to improve editing specificity, particularly in experiments aimed at dissecting the functional roles of specific mtDNA-encoded genes. Second, although we observed reduced expression of certain mitochondrially encoded proteins in some mt-Rnr1 mutant mice, there was no consistent inverse correlation between heteroplasmy and protein levels, as was observed with ATP levels. This inconsistency suggests the involvement of compensatory mechanisms at the transcriptional or translational level. Further investigations, including direct assessments of mitochondrial ribosomal activity, are thus required to better understand the extent of ribosomal dysfunction caused by mt-Rnr1 mutations.
In conclusion, despite these limitations, the development of Hifi-sTALED and αnHifi-sTALED represents a significant advancement in mitochondrial genome engineering. These optimized tools enable the efficient generation of animal models carrying mtDNA mutations, thereby offering new opportunities for the detailed study of mitochondrial diseases and their genetic underpinnings. Ultimately, these advancements hold strong potential for informing the development of novel therapeutic approaches targeting mitochondrial dysfunction.
Materials and methods
Animals
The Institutional Animal Care and Use Committee of Seoul National approved the experimental protocols involving mice. C57BL/6N mice were used as embryo donors, and ICR mice served as surrogate mothers. All mice were maintained in a specific pathogen-free facility under a 12-h light/dark cycle.
Cell lines
NIH3T3 cells were purchased from ATCC (CRL-1658). The culture medium for NIH3T3cells consists of Dulbecco’s modified Eagle’s medium (DMEM; Welgene) supplemented with 10% (v/v) bovine calf serum (Welgene) without antibiotics. Cells were incubated at 37°C with 5% CO2 and passed before approaching 80% confluency.
mRNA preparation
Plasmids encoding V28R-sTALED, Hifi-sTALED, and αnHifi-sTALED were used as templates for DNA polymerase chain reaction (PCR). Q5 High-Fidelity DNA Polymerase (New England Biolabs) was used with the following primers: F: 5′-CATCAATGGGCGTGGATAG-3′ and R: 5′-GACACCTACTCAGACAATGC-3′. In vitro transcription from the template DNA to mRNA was performed using the mMESSAGE mMACHINE T7 Ultra Kit (Ambion) and purified with the MEGAclear Kit (Ambion). Experiment procedures were performed with the manufacturer’s instructions.
Microinjection of mouse zygotes
Preparation of mouse zygotes for microinjection involved the following steps. First, 6- to 8-week-old female C57BL/6N mice were superovulated via an intraperitoneal injection of HyperOva (CARD, KYD-010-EX x5) and human chorionic gonadotropin (hCG; Sigma-Aldrich, CG10-1vl) at a 48-h interval. Then, these mice were mated with 10- to 26-week-old C57BL/6N male mice. One-cell mouse embryos were collected from the oviducts and placed in the M2 medium (Sigma-Aldrich, M7167). Cumulus cells were removed from zygotes by exposing them to 0.1% hyaluronidase (Sigma-Aldrich, H3884) in PBS and used for microinjections. The microinjection mixture was prepared with each split of V28R-, Hifi-, and αnHifi-sTALED at a concentration of 500 ng/μL (L/R each) and diluted in diethylpyrocarbonate (DEPC)-treated injection buffer (0.25 mM ethylenediaminetetraacetic acid [EDTA], 10 mM Tris; pH 7.4). Nikon ECLIPSE Ti micromanipulator and FemtoJet 4i microinjector (Eppendorf) were used for injection. After microinjection, zygotes were incubated in potassium simplex optimized medium (KSOM; MR-121-D, Millipore) at 37°C in a humidified atmosphere containing 5% CO2. Following incubation, two-cell stage embryos were transplanted into the oviducts of 0.5-day-post-coitus pseudo-pregnant surrogated mothers to obtain offspring. For in vitro analysis, untreated and microinjected zygotes were cultured in KSOM for 4 days to obtain blastocysts.
Genotyping and targeted deep sequencing
For preparation of DNA sample, blastocyst-stage embryos and tissues from mice were lysed in lysis buffer (25 mM sodium hydroxide [NaOH], 0.2 mM EDTA; pH 10) at 95°C for 20 min, and pH adjustment was used with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, free acids without pH adjustment) at a final concentration of 50 mM.
Targeted deep sequencing was conducted with sample lysate, nested first and second PCR were performed to create a high-throughput sequencing library, and index sequences are attached with third PCR. Q5 DNA polymerase (New England Biolabs) was used for PCR. Primers are listed on Supplementary Table. The library was subjected to paired-end read sequencing using MiniSeq (ILLUMINA). CRISPR RGEN Tools (http://www.rgenome.net/) was used for analyzing.
Whole mtDNA sequencing
DNA samples from tissues are obtained with DNeasy Blood & Tissue Kit (QIAGEN). For mtDNA amplification, PrimeSTAR GXL polymerase (Takara) was used and purified with Expin PCR SV (GeneAll) kit. Experiments were performed with the manufacturer’s instructions. Next-generation sequencing (NGS) library for MiSeq Sequencing System (Illumina) was prepared with Illumina DNA Prep kit and DNA CD Indexes. Experiment procedures were performed with the manufacturer’s instructions.
Measurement of ATP concentration
ATP concentrations in mutant NIH3T3 cell lines and mouse quadriceps were analyzed using the ATP Assay Kit (Abcam). All experimental procedures were performed according to the manufacturer’s instructions. To briefly explain the process, 10 mg of quadriceps tissue was homogenized in ATP assay buffer and centrifuged at 13,000×g for 5 min at 4°C. The supernatants were then diluted and mixed with reaction buffers in a 96-well plate (SPL, Seoul, Korea). After incubation at room temperature for 30 min in the dark, optical density (OD) was measured at 570 nm using a microplate reader.
Western blotting
First, mitochondria were isolated from the quadriceps muscles of wild-type and mutant mice using the Mitochondrial Isolation Kit (Abcam, ab110168), following the manufacturer’s protocol. Mitochondrial protein concentrations were determined using the Bradford protein assay (Bio-Rad). Equal amounts of total protein (5 μg per sample) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) 12% Premade gels (Bio-Rad, BR4561045) and transferred onto nitrocellulose membranes (Bio-Rad).
The membranes were probed with the following primary antibodies: Anti-Mouse MT-ND5 Antibody (LSBio, LS-C409671-20), MT-ATP6 Antibody (Cell Signaling Technology, #70262), COX1/MT-CO1 Antibody (CST, #62101), Cytochrome b (E7F8Z) Rabbit mAb (CST, #54618), and Anti-TOMM20 antibody (Abcam, ab186735).
After primary incubation, membranes were treated with HRP-conjugated secondary antibodies: Goat Anti-Rabbit IgG H&L (HRP) (Abcam, ab6721).
Protein bands were visualized using the Clarity Western ECL Substrate (Bio-Rad), and signals were detected with the iBright CL750 Imaging System. Band intensities were quantified by densitometry using ImageJ software (NIH) and normalized to TOMM20.
Oxygen consumption rates
Oxygen consumption rates were measured using the Seahorse XF HS Mini Analyzer (Agilent, Santa Clara, CA, USA) according to the manufacturer’s instructions. Eighty microliters of suspended cells (2 × 105 cells/mL) were seeded into Seahorse XF HS Miniplates (Agilent) 16 h prior to measurement. The analysis was conducted in Seahorse XF DMEM (pH 7.4) supplemented with 25 mM glucose and 1 mM sodium pyruvate (Agilent). Mitochondrial stress tests were performed using 1.5 mM oligomycin, 2 mM FCCP, and 0.5 mM rotenone plus antimycin A, as provided in the Seahorse XF Cell Mito Stress Test Kit (Agilent).
Data availability
Supplementary information is available in the online version of the paper. The high-throughput sequencing data from this study have been deposited in the NCBI Sequence Read Archive (SRA) database under accession code PRJNA1297430.
Acknowledgments
This work was supported by the National Research Foundation of Korea grants funded by the Korea government (MSIT) (RS-2024-00334994, RS-2023-00261905, RS-2024-00441068, RS-2024-00408822, RS-2025-02303431) and the Korea Institute for Advancement of Technology grants funded by the Korea government (MSIT) (P0028364).
Author contributions
S.H., and H.L. designed the research. S.H. and S.P.K. performed and analyzed the main experiments. S.Kim., S.K.K., S.H.C., S.B.L., J.K., Y.O., H.C., J.B., S.J., J.P., K.K., C.G.C., S.-J.P., D.K., and L.K.K. performed the experiments. H.L. and J.K.S. supervised the research. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2025.102678.
Contributor Information
Je Kyung Seong, Email: snumouse@snu.ac.kr.
Hyunji Lee, Email: hjlee102@korea.ac.kr.
Supplemental information
References
- 1.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., Hsu F., Radey M.C., Peterson S.B., Mootha V.K., et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho S.I., Lee S., Mok Y.G., Lim K., Lee J., Lee J.M., Chung E., Kim J.S. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell. 2022;185:1764–1776.e12. doi: 10.1016/j.cell.2022.03.039. [DOI] [PubMed] [Google Scholar]
- 5.Lee H., Lee S., Baek G., Kim A., Kang B.C., Seo H., Kim J.S. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat. Commun. 2021;12:1190. doi: 10.1038/s41467-021-21464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho S.I., Lim K., Hong S., Lee J., Kim A., Lim C.J., Ryou S., Lee J.M., Mok Y.G., Chung E., et al. Engineering TALE-linked deaminases to facilitate precision adenine base editing in mitochondrial DNA. Cell. 2024;187:95–109.e26. doi: 10.1016/j.cell.2023.11.035. [DOI] [PubMed] [Google Scholar]
- 7.Mok B.Y., Kotrys A.V., Raguram A., Huang T.P., Mootha V.K., Liu D.R. CRISPR-free base editors with enhanced activity and expanded targeting scope in mitochondrial and nuclear DNA. Nat. Biotechnol. 2022;40:1378–1387. doi: 10.1038/s41587-022-01256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Y., Jin M., Huang S., Yao F., Ren N., Xu K., Li S., Gao P., Zhou Y., Chen Y., et al. Enhanced C-To-T and A-To-G Base Editing in Mitochondrial DNA with Engineered DdCBE and TALED. Adv. Sci. 2024;11 doi: 10.1002/advs.202304113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S., Lee H., Baek G., Kim J.S. Precision mitochondrial DNA editing with high-fidelity DddA-derived base editors. Nat. Biotechnol. 2023;41:378–386. doi: 10.1038/s41587-022-01486-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo S.R., Simone B.W., Clark K.J., Devaux P., Ekker S.C. Unconstrained Precision Mitochondrial Genome Editing with alphaDdCBEs. Hum. Gene Ther. 2024;35:798–813. doi: 10.1089/hum.2024.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotrys A.V., Durham T.J., Guo X.A., Vantaku V.R., Parangi S., Mootha V.K. Single-cell analysis reveals context-dependent, cell-level selection of mtDNA. Nature. 2024;629:458–466. doi: 10.1038/s41586-024-07332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lake N.J., Ma K., Liu W., Battle S.L., Laricchia K.M., Tiao G., Puiu D., Ng K.K., Cohen J., Compton A.G., et al. Quantifying constraint in the human mitochondrial genome. Nature. 2024;635:390–397. doi: 10.1038/s41586-024-08048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zong Y., Li H., Liao P., Chen L., Pan Y., Zheng Y., Zhang C., Liu D., Zheng M., Gao J. Mitochondrial dysfunction: mechanisms and advances in therapy. Signal Transduction Targeted Ther. 2024;9:124. doi: 10.1038/s41392-024-01839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen H., Deng H., Li B., Chen J., Zhu J., Zhang X., Yoshida S., Zhou Y. Mitochondrial diseases: from molecular mechanisms to therapeutic advances. Signal Transduction Targeted Ther. 2025;10 doi: 10.1038/s41392-024-02044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturm G., Karan K.R., Monzel A.S., Santhanam B., Taivassalo T., Bris C., Ware S.A., Cross M., Towheed A., Higgins-Chen A., et al. OxPhos defects cause hypermetabolism and reduce lifespan in cells and in patients with mitochondrial diseases. Commun. Biol. 2023;6:22. doi: 10.1038/s42003-022-04303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Hong M., Luan C., Yuan M., Wang Y., Guo X., Fang Y., Huang H., Dong X., Gao H., et al. Efficient mitochondrial A-to-G base editors for the generation of mitochondrial disease models. Nat. Biotechnol. 2025 doi: 10.1038/s41587-025-02685-x. [DOI] [PubMed] [Google Scholar]
- 17.Fan Y., Xu W., Gao B.Q., Qin H., Wu X., Wei J., Ni Q., Zhou L., Xiang J., Wu J., et al. Leveraging base excision repair for efficient adenine base editing of mitochondrial DNA. Nat. Biotechnol. 2025 doi: 10.1038/s41587-025-02608-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary information is available in the online version of the paper. The high-throughput sequencing data from this study have been deposited in the NCBI Sequence Read Archive (SRA) database under accession code PRJNA1297430.