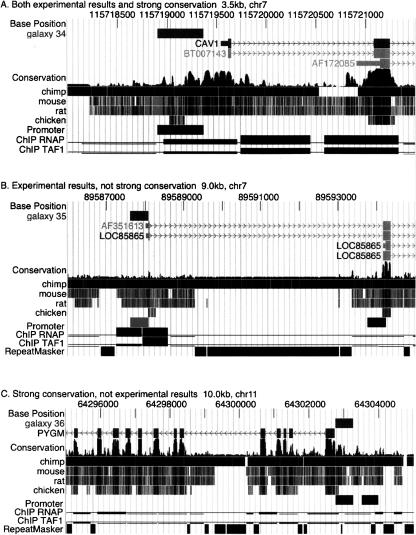
Figure 3.
Examples of promoters characterized by binding of the transcription initiation complex and/or high conservation. Images from the UCSC Genome Browser generated via Galaxy illustrate (A) a promoter that has strong conservation (indicative of purifying selection) and biochemical evidence of binding by RNA polymerase II and TAF1, (B) a promoter that is poorly conserved but is strongly bound by RNA polymerase II and TAF1, and (C) a strongly conserved promoter that is not bound by the transcription initiation machinery in the cells tested. The track labeled “galaxy” is the custom track automatically generated by Galaxy for each query number (34, 35, and 36). Genes are labeled and have exons as boxes and introns as lines with arrowheads pointing in the direction of transcription. “Conservation” is the phastCons track followed by positions of aligning DNA in homologous regions of other species. The positions of promoters are shown as rectangles. The results of chromatin immunoprecipitations (ChIP data) are plotted as the negative log of the p-value, ranging in the vertical direction from 0 to 10.0 and with a continuous thin line placed at the threshold of 2.0. Positions of repeats identified by RepeatMasker (A.F.A. Smit and P. Green, unpubl., http://ftp.genome.washington.edu/RM/RepeatMasker.html) are shown as black rectangles in panels B and C.