Abstract
Aminoacyl-tRNA synthetase-containing complexes have been identified in different eukaryotes, and their existence has also been suggested in some Archaea. To investigate interactions involving aminoacyl-tRNA synthetases in Archaea, we undertook a yeast two-hybrid screen for interactions between Methanothermobacter thermautotrophicus proteins using prolyl-tRNA synthetase (ProRS) as the bait. Interacting proteins identified included components of methanogenesis, protein-modifying factors, and leucyl-tRNA synthetase (LeuRS). The association of ProRS with LeuRS was confirmed in vitro by native gel electrophoresis and size exclusion chromatography. Determination of the steady-state kinetics of tRNAPro charging showed that the catalytic efficiency (kcat/Km) of ProRS increased 5-fold in the complex with LeuRS compared with the free enzyme, whereas the Km for proline was unchanged. No significant changes in the steady-state kinetics of LeuRS aminoacylation were observed upon the addition of ProRS. These findings indicate that ProRS and LeuRS associate in M. thermautotrophicus and suggest that this interaction contributes to translational fidelity by enhancing tRNA aminoacylation by ProRS.
Aminoacyl-tRNA synthetases (aaRSs)1 are essential components of the translation process. Their cellular role is to ensure that individual tRNAs are attached to their cognate amino acid. Each aaRS specifically binds to a defined set of tRNAs and catalyzes the attachment of the amino acids to the tRNAs. Once synthesized, the aminoacyl-tRNAs function as substrates for ribosomal protein synthesis, thereby ensuring correct translation of the genetic code (1). In bacteria, aaRSs typically perform their role as individual enzymes, found either as monomers, homodimers, or homo- or heterotetramers. However, in eukaryotes several aminoacyl-tRNA synthetases exist in multienzyme complexes (2–4), and two different types have so far been found in mammalian cells. One is composed of only one aminoacyl-tRNA synthetase, valyl-tRNA synthetase, and EF-1H, the heavy form of translation elongation factor 1 (5). The complex is believed to contain seven subunits, two monomeric subunits of valyl-tRNA synthetase and the EF-1H subunits EF1-α, -β, -γ, and -δ in the molar ratio 2:1:1:1. The second complex described is considerably larger and includes nine aminoacyl-tRNA synthetase activities. The complex is composed of isoleucyl-, leucyl- (LeuRS), prolyl-(ProRS), methionyl-, glutaminyl-, glutamyl-, lysyl-, arginyl-, and aspartyl-tRNA synthetases. Whereas most of the aaRSs are present in the complex as monomers, data have indicated that lysyl-tRNA synthetase and aspartyl-tRNA synthetase exist as dimers (3, 6). In addition, the polypeptide carrying the ProRS activity is multifunctional in that the protein also comprises the catalytic domain and activity of glutamyl-tRNA synthetase (7). Three auxiliary proteins, p18, p38, and p43, are also part of the multisynthetase complex. Although the structural and functional significance of the complex still remains to be elucidated, it is known that N- and C-terminal extensions of the mammalian synthetases mediate association of the components. The accessory components p18, p38, and p43 assist complex formation and stability and promote tRNA binding by the complex (8–10). The heat shock protein Hsp90 has also been found to bind aaRSs of the complex (11), an interaction believed to facilitate assembly.
The only other multi-aaRS complex so far identified in eukaryotes was discovered in the yeast Saccharomyces cerevisiae. The complex consists of methionyl-tRNA synthetase, glutamyl-tRNA synthetase, and the nonsynthetase protein Arc1p, which has homology to the mammalian protein p43 (12, 13). The association with Arc1p was shown to increase the catalytic efficiency of the two synthetases and enhances nuclear export of tRNA. The bacterial homologue of Arc1p, Trbp111, was first found in the extreme thermophile Aquifex aeolicus and was shown to promote tRNA binding by aaRSs (14, 15). Factors unrelated to the translation machinery have also been found to associate with aaRSs. In one case, a two-hybrid screen revealed interaction between yeast seryl-tRNA synthetase and Pex21p, a protein involved in peroxisome biogenesis (16). In a similar screen, yeast tyrosyl-tRNA synthetase was isolated as a protein associating with Knr4p, a protein involved in regulation of cell wall assembly (17).
In Archaea, much less is known about aaRS complexes, and to date only two studies have reported their possible existence. Methanocaldococcus jannaschii ProRS was co-purified with the H2-forming N5-N10-methylene tetrahydromethanopterin dehydrogenase (HMD), a component of the methanogenesis pathway (18). ProRS and HMD were also used for co-immunoprecipitation experiments with recombinant proteins, leading to the proposal that HMD specifically interacts not only with ProRS but also with lysyl-tRNA synthetase and aspartyl-tRNA synthetase. The cellular role of the complex remains unclear, since the aaRS activities are unchanged upon complex formation. An aaRS complex has also been described in the extreme halophile Haloarcula marismortui, with many if not all of the aaRSs purified in one or possibly two large complexes (19). Further investigations of archaeal aaRS complexes are warranted, both to more clearly understand their role in these unusual organisms and also to better understand the role of such interactions in general. We have now used the yeast two-hybrid system to search for proteins interacting with ProRS in the archaeal methanogen Methanothermobacter thermautotrophicus (20). This identified a stable interaction between LeuRS and ProRS, which appears to specifically enhance tRNAPro aminoacylation.
EXPERIMENTAL PROCEDURES
Media, Strains, and Plasmid Construction
Cloning procedures and Escherichia coli media were prepared by standard methods. Yeast transformation was done according to Ref. 21. Yeast media were made according to the manual for the ProQuest two-hybrid system (Invitrogen) and as described (22). All primers were from Integrated DNA Technologies. The bait vector pDBLeu, prey vector pDEST22, and yeast host strain MaV203 (MATα leu2–3, 112, trp 1–901, his3Δ200, ade2–101, gal4Δ, gal80Δ, SPAL10::URA3, GAL1::lacZ, HIS3UAS GAL1::HIS3@LYS2, can1R, cyh2R) were from the ProQuest two-hybrid system (Invitrogen). Construction of yeast two-hybrid bait vector containing the M. thermautotrophicus proS gene was done as follows. The ProRS-encoding gene, (proS, MTH611), was isolated by PCR using genomic M. thermautotrophicus DNA as template, the primers 5′-GGTGTTGTCGACCATGCAGAAACCTATC-3′ and 5′-GGTGGTGTCGACTCAGCTAATATGTTC-3′ flanked by SalI sites and Pfu DNA polymerase (Stratagene). The proS PCR product was cloned into PCR-Blunt II-TOPO vector (Invitrogen), sequenced, and subsequently subcloned into the yeast ProQuest two-hybrid bait vector pDBLeu using the SalI restriction sites. The sequence obtained from verifying the proS clones deviated from the published MTH611 sequence in that it contained an extra C at position 1412. The C insertion in the sequence gave rise to a frameshift that then encoded the last nine amino acids of ProRS as YRAYLARTY instead of GLTLPEHIS (Stehlin et al. (23) also reported LARTY as the five last amino acids).
N-terminally tagged His6 fusion derivatives of ProRS and LeuRS (MTH1508) were made by PCR amplification of the relevant genes using oligonucleotide primers containing sequences encoding six histidine residues located immediately after the start codons of the genes (see below). Appropriate restriction sites used for cloning into final expression vectors were also included. Templates were either genomic DNA or plasmids containing the relevant genes. The PCR products were cloned into PCR-Blunt II-TOPO vector (Invitrogen) and sequenced prior to cloning into the E. coli expression plasmid pET11a (Novagen). For the His6-ProRS construct, primers 5′-CATATGCATCACCATCACCATCACCAGAACCTATCAAA-3′ and 5′-TGATCATTAGCTAATATGTTC-3′-were used, and for His6-LeuRS, 5′CATATGCATCACCATCACCATCACGATATTGAAAGAAAATGG-3′ and 5′-TGATCATTATTCAAGGTATATGGCTGGCT-3′ were used. Cloning into pET11a was done by isolating the respective NdeI-BclI fragments and ligating them into NdeI-BamHI-digested pET11a.
Preparation of M. thermautotrophicus mRNA
Six cultures of M. thermautotrophicus were grown in 1.5-liter bioreactors at 55 °C in a minimal salts medium (24). A gas mixture of 89% H2, 11% CO2 was supplied to the cultures at a flow rate of 200 ml/min. The impellers on the fermentation vessels were spun at either 600 rpm to allow an optimum amount of hydrogen gas to dissolve into the medium or at 200 rpm to limit hydrogen dissolution. Eight RNA samples were purified from the six cultures under both high and low hydrogen conditions during early, middle, and late logarithmic growth phases. To extract the RNA, 10 ml of culture were withdrawn from the fermentation vessels into disposable Becton Dickinson syringes at each of the time points. The samples were then passed through reusable Gelman filter units containing Millipore nitrocellulose filters, 0.45-μm pore size. The filters were placed inside 2-ml screw cap vials containing 0.6 ml of 0.1-mm zirconia/silica beads; 0.6 ml of saturated phenol, pH 4.3; and 0.6 ml of 1% SDS, 0.1 m sodium acetate, pH 5.2. The tubes were agitated inside a Mini-BeadBeater-8 cell disrupter (BioSpec Products, Inc.) for 5 min at 3,200 rpm. After centrifugation, the aqueous phase was removed from each of the samples and extracted again with saturated phenol, pH 4.3. Nucleic acids were precipitated overnight with isopropyl alcohol and 0.3 m sodium acetate, pH 5.2. The pellets were washed with 75% ethanol, allowed to air-dry, and then resuspended in 50 μl of RNase-free distilled H2O. The samples were treated with DNase (amplification grade; Invitrogen) according to the supplier’s instructions. RNA was further purified using a Qiagen RNeasy kit according to the manufacturer’s instructions. The concentrations of the samples were determined spectrophotometrically, and Northern blots were performed to check for degradation. 10 μg of total RNA from eight different samples were pooled (80 μg total) to provide templates for cDNA synthesis.
Construction of M. thermautotrophicus cDNA-based Yeast Two-hybrid Library
cDNA library was generated by Christian Gruber and Mark Smith (Invitrogen) using random priming of M. thermautotrophicus total RNA. The cDNA was directionally cloned into the pDEST-22 vector. The resulting cDNA library contains 2.6 × 106 clones (representing >1000-fold coverage) with an average insert size of 0.4–2 kb.
Yeast Two-hybrid Screen
M. thermautotrophicus proS cloned in the yeast bait vector pDBLeu (ProQuest two-hybrid system; Invitrogen) was used to co-transform the yeast strain MaV203 with an M. thermautotrophicus cDNA library cloned into the prey vector pDEST22. Potential positive clones were selected by plating transformants on SC-Leu-Trp-His + 3-aminotriazole (3-AT; 10 mm) and incubating the plates at 30 °C for 3–10 days including replica cleaning as described (ProQuest two-hybrid system). The transformation plates were then replica-plated onto SC-Leu-Trp + 3-AT (10 and 25 mm) and incubated as above. Transformants showing consistent growth were streaked for single colonies, and from each transformant four colonies were retested for growth on SC-Leu-Trp-His + 10, 25, and 35 mm 3-AT and also tested for phenotypes of the other reporter genes (i.e. growth on SC-Leu-Trp-Ura and no growth on SC-Leu-Trp + 0.2% 5-fluoroorotic acid). A total of 2.8 × 105 transformants were screened, of which 90 were identified as positive interacting clones. Isolation of positive clones for sequencing was done by growing the co-transformants in SC-Trp followed by plating on SC-Trp in order to isolate colonies harboring only the prey vector. These colonies were tested for their inability to grow on media lacking leucine before prey plasmids were isolated from the cultures (25) and inserts were sequenced. Prey vector cDNA inserts were sequenced using an oligonucleotide matching the part of the prey vector that reads into the 5′-end of the insert. In a few cases, oligonucleotides that surround the insert were used, so both the sequences in the 5′-end and the 3′-end of inserts could be determined.
Protein Production and Purification
Production of His6-tagged ProRS and LeuRS was done by transforming BL21 (DE3) RP or RIL strains (Stratagene) with pET11 containing the relevant inserts and growing the resulting strains using the Overnight Express™ Auto-induction System 1 (Novagen) according to the manufacturer’s instructions. For His6-LeuRS, cell-free extract was prepared by sonication of the E. coli cells in lysis buffer (50 mm NaH2PO4, 300 mm NaCl) containing protease inhibitor mixture tablet (Complete Mini, EDTA-free; Roche Applied Science) followed by centrifugation at 27,000 rpm for 20 min. To minimize contamination with E. coli proteins, a flocculation step at 55 °C for 10 min was included prior to ultracentrifugation at 40,000 rpm for 1 h. The supernatant was then applied to a Ni2+-nitrilotriacetic acid Superflow column (Qiagen) equilibrated in lysis buffer and extensively washed in the same buffer containing 10 mm imidazole. His6-LeuRS was eluted in the same buffer containing 250 mm imidazole. Fractions containing His6-LeuRS (judged by Coomassie Brilliant Blue staining after SDS-PAGE) were pooled, and buffer was exchanged to buffer A (50 mm Hepes, pH 7.2, 25 mm KCl, 10 mm MgCl2, 5 mm dithiothreitol, 10% glycerol) using a HiPrep 26/10 desalting column (GE Healthcare). Samples were then concentrated and further purified to >95% purity by ultrafiltration (Amicon Ultra-15; 30-kDa cut-off). Aliquots were stored at −80 °C. Protein extract used to purify His6-ProRS was handled as described for LeuRS, except that the flocculation step was omitted. Fractions eluted and pooled from the Ni2+-nitrilotriacetic acid Superflow column were diluted 5-fold in H2O and subjected to ultrafiltration as above. Samples were then diluted 5-fold in buffer A and applied to a Resource Q column (GE Healthcare) and eluted with a NaCl gradient (0–500 mm) in the same buffer. Fractions containing ProRS (judged by Coomassie Brilliant Blue staining after SDS-PAGE) were pooled and resubjected to ultrafiltration, aliquoted, and stored at −80 °C. ProRS prepared in this way was judged to be >95% pure. The concentrations of LeuRS and ProRS were determined by active site titration as previously described (26), and the reaction was performed for 5 min.
Native Gel Electrophoresis and Immunoblotting
Attempts to visualize ProRS·LeuRS complexes by native PAGE were done by incubating a 1–2 μm concentration of each protein together in 10 mm Tris-HCl, pH 7.0, 5 mm MgCl2 at 4 °C for 15 min (30-μl sample volume). Glycerol was added to the samples to a final concentration of 5% prior to loading on a 12% native polyacrylamide gel and run at 100 V for 16 h at 4 °C in native running buffer (25 mm Tris, 0.2 m glycine, pH 8.5). Attempts to increase ProRS·LeuRS complex formation were made by incubating proteins at room temperature for 15–30 min combined with running gels at room temperature for 6 h at 100 V or for 16 h at 35 V. These changes did not result in increased visualization of protein-protein complex formation between ProRS and LeuRS. Western blotting was done in order to visualize the high molecular weight complex, as well as His6-LeuRS and His6-ProRS alone, by using antibodies against the epitope tag (tetra-His antibody; Qiagen), anti-mouse Ig horseradish peroxidase as second antibody (GE Healthcare), ECL plus Western blotting detection system, and hyperfilm ECL (GE Healthcare).
Aminoacylation Assays
l-[U-14C]leucine (331 mCi/mmol), l-[U-14C]proline (276.5 mCi/mmol), and l-[2,3,4,5-3H]proline (85.0 Ci/mmol) were all from PerkinElmer Life Sciences. The gene encoding M. thermautotrophicus tRNALeu (GAG anticodon) was cloned into pUC18, whereas the corresponding tRNAPro was a gift from D. Söll (Yale University). Synthesis and purification of the corresponding in vitro transcribed tRNAs was performed using standard procedures (27). All aminoacylations were performed at 50 °C as follows. A prereaction mixture was first prepared containing 100 mm Hepes (pH 7.5), 125 mm dipotassium glutarate, 250 mm KCl, 10 mm MgCl2, 10 mm dithiothreitol, 6 μg/μl M. thermautotrophicus total tRNA (prepared as per Ref. 28), or in vitro transcribed tRNA and aaRSs at the concentrations indicated for specific experiments. The entire mixture was incubated for 20 min at room temperature, the appropriate radiolabeled amino acid was added, and the temperature was shifted to 50 °C. After 1 min, the reaction was started by the addition of 5 mm ATP. Aliquots were removed periodically and spotted onto 3MM filter disks presoaked in 5% trichloroacetic acid (w/v) and then washed, and radioactivity was counted as described previously (29). For Pro Km determination, l-[3H]Pro was added at concentrations varying between 0.2 and 5 times Km. For tRNAPro Km determination, tRNA was added at concentrations varying between 0.2 and 5 times the Km. Due to the relatively low activity of in vitro transcribed tRNALeu, saturation conditions could not be achieved, and kcat/Km was estimated directly (see “Results” for details).
Size Exclusion Chromatography of aaRSs
Size exclusion chromatography was performed using a Superose 12 column (GE Healthcare) pre-equilibrated in low salt (50 mm Hepes (pH 7.5), 25 mm KCl, 10 mm MgCl2, 10 mm dithiothreitol) or high salt buffer (100 mm Hepes (pH 7.5), 125 mm dipotassium glutarate, 250 mm KCl, 10 mm MgCl2, 10 mm dithiothreitol). Both columns were calibrated using gel filtration standard (Bio-Rad). Samples were prepared in the same buffer and contained 1.4 μm His6-ProRS, 1.4 μm His6-LeuRS (both 100-μ l injected sample volume), or a mixture of 2 μm His6-ProRS and 1.8 His6-LeuRS μm (200-μl injected sample volume).
RESULTS
Identification of ProRS-interacting Proteins
The bait vector, pDBLeu, harboring the M. thermautotrophicus ProRS-encoding gene was used in the two-hybrid screen with a cDNA library cloned into the prey vector pDEST22. A total of 2.8 × 105 transformants were screened for protein-protein interacting phenotypes. Of those, 525 potential positive clones were retested for the protein-protein interacting phenotype, and 90 clones were selected for plasmid isolation and sequencing. Five clones, each identified once in the screen, contain genes involved in different steps in the methanogenesis pathway (MTH1159, MTH1160, MTH1163, MTH1300, and MTH1878; Table I). Two of four protein-modifying genes identified (MTH785 and MTH1623) appeared once, whereas the other two appeared eight times in total (MTH357 and MTH412). LeuRS (MTH1508) was identified once, and other proteins that are all subunits of protein complexes appeared either once (MTH736 and MTH802) or twice (MTH957 and MTH674).
Table I.
Proteins identified as interacting with M. thermautotrophicus ProRS
| Function | Open reading framea | Descriptionb |
|---|---|---|
| Translation | MTH1508 | Leucyl-tRNA synthetase |
| Protein modification | MTH357 | Transglutaminase |
| MTH412 | Transglutaminase | |
| MTH785 | La protease | |
| MTH1623 | Oligosaccharyl transferase STT3 | |
| Methanogenesis | MTH1159 | N5-methyltetrahydromethanopterin:CoM methyltransferase, α-subunit |
| MTH1160 | N5-methyltetrahydromethanopterin:CoM methyltransferase, β-subunit | |
| MTH1163 | N5-methyltetrahydromethanopterin:CoM methyltransferase, E-subunit | |
| MTH1300 | Co-F420-reducing hydrogenase, a-subunit | |
| MTH1878 | CoB-CoM heterodisulfide reductase, C-subunit | |
| Other | MTH736 | Carbomoyl phosphate synthase large subunit |
| MTH802 | Aspartokinase II α-subunit | |
| MTH957 | ATP synthase | |
| MTH674 | Unknown function |
ORFs are numbered according to the annotated genome sequence of M. thermautotrophicus (20).
Predicted functions of the corresponding proteins are taken from the TIGR Comprehensive Microbial Resource (available on the World Wide Web at www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl).
Potential ProRS-associating proteins identified from the two-hybrid screen were sorted into four subgroups (Table I). The first contained protein-modifying enzymes known to bind a large variety of proteins in the cell. The most abundant of these was a transglutaminase-like superfamily domain protein (MTH412) and its homologue (MTH357), both of which have been annotated as proteins involved in protein degradation. In addition, a putative La protease (MTH785) and an oligosaccharyl transferase STT3 subunit-related protein (MTH1623) were found, both of which act in modification or degradation of other proteins. All of the members of this group generally bind to many different proteins in the cell and thus are more likely to act as expected false positive clones rather than being proteins that specifically bind ProRS. The second major grouping contained three subunits from different multimeric proteins (MTH736, -802, and -957) and one protein of unknown function (MTH674). As with the first grouping, the finding that most of these proteins are normally expected to associate with other proteins suggests that they may also be false positive clones. The other large group of proteins identified in this study contains components of the methanogenesis pathway (MTH1159, -1160, -1163, -1300, and -1878). All of these proteins are known to be subunits of larger multimeric proteins and thus might be expected to readily associate with other proteins, perhaps as components of higher order complexes (e.g. see Ref. 30). The only component of protein synthesis identified from the two-hybrid screen was LeuRS, a monomeric aaRS. Contact between ProRS and LeuRS has previously been observed in the human multi-aaRS complex (reviewed in Refs. 3 and 6), and the functional consequences of this interaction in Archaea were further characterized (see below). In light of the potential for nonspecific complex formation, the ability of proteins other than LeuRS to interact with ProRS was not further investigated.
Association of ProRS with LeuRS
A gene fragment encoding the C-terminal 471 amino acids (residues 467–938) of LeuRS was isolated as a ProRS-interacting clone in the two-hybrid screen (Fig. 1 and Table II). In order to further investigate complex formation between ProRS and full-length LeuRS, both proteins were produced and purified heterologously as His6-amino-terminal fusion proteins. Complex formation was first monitored by native gel electrophoresis followed by immunoblotting with His6-specific antibodies (Fig. 2). LeuRS and ProRS are easily detected and well resolved under the conditions employed here (Fig. 2, lanes 1 and 2, respectively). When ProRS and LeuRS are incubated together prior to analysis, an additional species is observed with a slower mobility than either of the individual proteins (Fig. 2, lanes 3–5), suggesting that a complex is formed between the two proteins. Attempts to further increase the level of ProRS·LeuRS complexes by using a variety of different conditions were unsuccessful (data not shown). The low proportion of total proteins visible by gel electrophoresis in the ProRS·LeuRS complex may be a consequence of the relatively high salt concentrations required for optimal association (see below), conditions incompatible with native gel electrophoresis.
Fig. 1. Yeast two-hybrid interaction between LeuRS and ProRS.
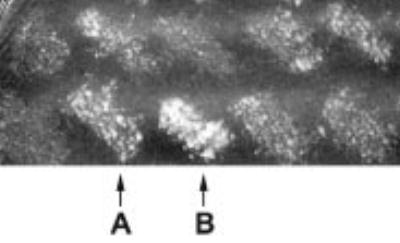
Shown is the growth phenotype on selective media (SC lacking leucine and tryptophan, supplemented with 35 mm 3-AT) for interactions between ProRS (encoded in the bait vector pDBleu-proS) and library inserts (encoded in the prey vector pDEST). A portion of the secondary screening is shown (see “Experimental Procedures” for details) including interactions with ProRS subsequently identified as glutaminase (A) and LeuRS (B).
Table II. Yeast two-hybrid interaction between LeuRS and ProRS.
Growth phenotypes on selective media for interactions between reference proteins (A–D are controls (ProQuest two-hybrid system; Invitrogen)) and between ProRS (encoded in the bait vector pDBleu-proS) and library inserts (encoded in the prey vector pDEST) are shown. Growth of transformed MaV203 after 48 h at 30 °C on SC lacking leucine and tryptophan supplemented as indicated is shown. −, no growth; +, weak growth; ++, moderate growth; +++, strong growth.
| Plasmids | −Ura | −His, +25 mm 3-AT | +0.2% 5-fluoroorotic acid | Description |
|---|---|---|---|---|
| A controls | − | - | ++ | No Interaction standard |
| B controls | +/− | + | + | Weak Interaction standard |
| C controls | ++ | + | +/− | Moderate interaction standard |
| D controls | +++ | +++ | ++ | Strong interaction standard |
| pDBleu-proS/pDEST-I149 | − | + | +++ | Glutaminase insert in prey vector |
| pDBleu-proS/pDEST-I150 | +/− | +++ | +++ | LeuRS insert in prey vector |
| pDBleu-proS/+ pPC86 | − | +/− | +++ | Empty prey vector control |
Fig. 2. Association of LeuRS with ProRS.
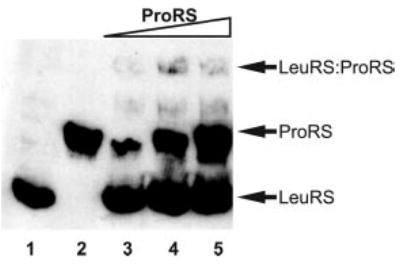
Native gel electrophoresis of His6-LeuRS and His6-ProRS visualized by immunoblotting using antibodies against the His6 epitope. LeuRS alone (1.6 μm; lane 1), ProRS alone (1.8 μm; lane 2), LeuRS (1.6 μm), and ProRS combined (0.6/1.2/1.8 μm; lanes 3–5) are shown.
The association of ProRS with LeuRS was further investigated by size exclusion chromatography using a Superose 12 column. Initial experiments were performed using the same buffer conditions as described above for native gel electrophoresis. This led to the appearance of an additional minor species (<1% of loaded protein) with a predicted molecular weight consistent with a complex stoichiometry of 2 ProRS:2 LeuRS (Fig. 3A). The presence of both LeuRS and ProRS in this higher molecular weight fraction was confirmed by detection of the corresponding aminoacylation activities using total tRNA from M. thermautotrophicus as substrate. In an attempt to increase the proportion of proteins in the complex, buffer composition was adjusted to more closely reflect the intracellular environment of M. thermautotrophicus (addition of 125 mm dipotassium glutarate and 250 mm KCl (31, 32)). Under these conditions, the level of complex increased significantly and accounted for about 30% of the total protein (Fig. 3B). The exact stoichiometry of the complex could not be readily deduced from its retention time, because both the standards and the individual proteins behaved atypically at elevated salt concentrations (Fig. 3B, inset).
Fig. 3. Gel filtration chromatography of LeuRS and ProRS.
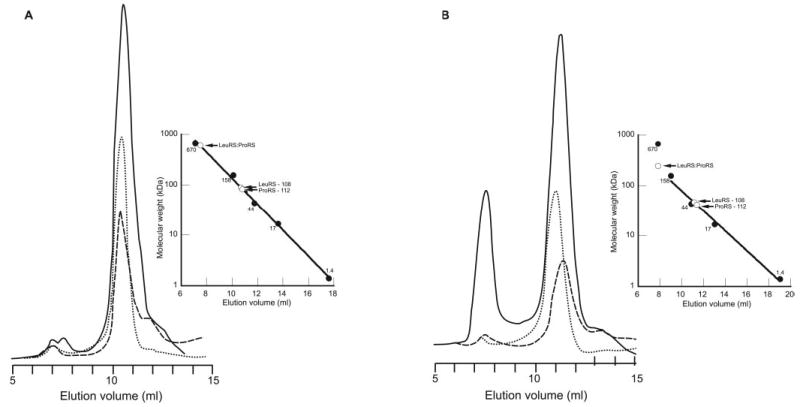
LeuRS and ProRS were applied either alone or combined after preincubation to a Superose 12 column equilibrated in low (A) or high (B) salt buffer. Sample chromatograms (A280 nm) for LeuRS (dotted line), ProRS (dashed line), and LeuRS·ProRS (solid line) at low salt (V0 = 7.0 ml) and high salt (V0 = 7.3 ml) are shown. Calibration curves (inset) show the elution positions of both individual and combined proteins.
Kinetics of Aminoacylation by the ProRS·LeuRS Complex
Potential functional effects of the association between ProRS and LeuRS were investigated in the aminoacylation reaction using high salt conditions similar to those described above. In order to ensure that the majority of the monitored enzyme was in the complex, a 10-fold excess of the other protein was employed. Experiments employing a short preincubation period indicated that the addition of LeuRS initially suppressed aminoacylation by ProRS, whereas extended preincubation led to an increase in the rate of aminoacylation (Fig. 4). Determination of steady-state kinetic parameters for aminoacylation indicated that complex formation both decreased the Km for tRNAPro and increased the kcat of ProRS (Table III), leading to a 5-fold increase in kcat/Km for tRNAPro (Table III). Kinetic parameters for proline showed a comparable change in kcat, whereas Km was unchanged. Nonspecific effects of the addition of excess partner protein were excluded by the addition of similar quantities of BSA, which moderately enhanced aminoacylation by both ProRS and ProRS·LeuRS to the same degree (Table III). The further addition of BSA beyond the concentration initially employed did not result in any further changes in aminoacylation (data not shown). The possible enhancement of aminoacylation by LeuRS in the presence of excess ProRS was also investigated, but no significant difference was observed compared with the activity of LeuRS alone (Table IV).
Fig. 4. Aminoacylation of tRNAPro.
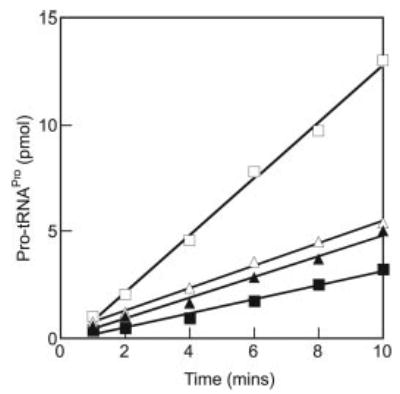
In vitro transcribed tRNAPro (16 μm) was aminoacylated under standard conditions (9-μl samples) in the presence of ProRS (40 nm) either with (open triangles) or without (filled triangles) preincubation for 20 min at room temperature. The reactions were also performed with ProRS (40 nm) and LeuRS (400 nm) either with (open squares) or without (filled squares) preincubation.
Table III.
Steady-state aminoacylation kinetics of M. thermautotrophicus ProRS
| Enzymea | Additionsb | Km Pro | Km tRNAPro | kcat | kcat/Km tRNA (relative) |
|---|---|---|---|---|---|
| μm | μm | s−1 | |||
| ProRS | None | NDc | 4.1 ± 0.9 | 1.8 ± 0.2 | 1 |
| ProRS | LeuRS | ND | 2.2 ± 0.6 | 5.1 ± 0.4 | 5 |
| ProRS | BSA | 67 ± 10 | 2.4 ± 0.4 | 2.9 ± 0.1 | 3 |
| ProRS | LeuRS, BSA | 61 ± 17 | 1.4 ± 0.1 | 6.9 ± 0.2 | 12 |
Enzymes were added at a final concentration of 40 nm.
Other components at 400 nm each.
ND, not determined.
Table IV.
Steady-state aminoacylation kinetics of M. thermautotrophicus LeuRS
| Enzymea | Additionsb | kcat/Km Leu | kcat/Km tRNALeu |
|---|---|---|---|
| s−1 μm−1 | |||
| LeuRS | BSA | 0.35 ± 0.07 | 0.75 ± 0.08 |
| LeuRS | ProRS, BSA | 0.37 ± 0.09 | 0.75 ± 0.03 |
Enzymes were added at a final concentration of 40 nm.
Other components at 400 nm each. Due to the high Km compared to practical tRNA concentrations ([S] ≪ Km), kcat/Km was directly estimated from the equation v = kcat / Km ([E] [S]).
DISCUSSION
One of the major groups of proteins found to interact with ProRS in the two-hybrid assay acts in the methanogenesis pathway. There are seven steps in the H2-dependent reduction of CO2 to CH4 and an eighth energy conservation step. Proteins were identified in the ProRS-mediated two-hybrid screens that are parts of the complexes carrying out three of the eight steps. The two-hybrid screen did not identify the protein HMD, which in M. jannaschii co-purified with ProRS (18). HMD catalyzes the conversion of methenyl-H4MPT to methylene-H4MPT, the fourth step in the reduction of CO2 to CH4 in M. thermautotrophicus (33). HMD not only co-eluted from an affinity resin with M. jannaschii His-tagged ProRS but also with Homo sapiens His-tagged ProRS and lysyl-tRNA synthetase. This ability of HMD to associate with both homologous and heterologous synthetases, together with its tRNA binding capacity, suggests that it may function as a tRNA-interacting factor, as also recently proposed for p43/Arc1p in eukaryotes (34). Whereas HMD itself or known similar factors were not detected here, ProRS did identify a subunit of a protein that under certain conditions provides reduced coenzyme F420 for methylene-H4MPT formation. The independent identification in this study of interactions between ProRS and enzymes contributing to the same step in methanogenesis as HMD supports the proposed link between metabolism and decoding in Archaea (18), although the role of such a connection remains unknown.
In addition to the interactions with HMD described above, aaRS complexes were also previously reported from the halophilic archaeon H. marismortui (19). One, or possibly two, stable complexes containing proteins both with and without aminoacylation activity were identified, and it was suggested that the aaRSs in these complexes are kept together by hydrophobic interactions. Since the elevated salt concentrations observed in H. marismortui and, to a lesser extent, M. thermautotrophicus may not be beneficial for synthetase-tRNA interactions, the multisynthetase particle could provide the necessary environment for efficient aminoacylation of tRNA by aminoacyl-tRNA synthetases. This is indeed the case for the interaction between LeuRS and ProRS described here, which leads to improved aminoacylation by ProRS under moderately halophilic conditions in vitro. Preliminary affinity co-precipitation experiments using M. thermautotrophicus cell extracts suggest that in vivo LeuRS and ProRS interact as part of a larger complex that includes lysyl-tRNA synthetase, further supporting the notion that aaRS interactions provide an optimal environment for aminoacylation.2
Beyond the Archaea, LeuRS and ProRS have also been found in close proximity as components of the same subdomain of the mammalian multi-aaRS complex (35, 36). LeuRS and ProRS are believed to be anchored to the C-terminal end of the auxiliary protein p38 (37), which acts as a scaffold protein and is essential for complex assembly and stability (10, 37). Another auxiliary factor of the mammalian complex, p43, which is located in the core of the particle (4, 35), has also been implicated in complex assembly. p43 orthologs have been identified in yeast (Arc1p) and bacteria (Trbp111) (12, 14). This group of proteins preferentially binds tRNA and facilitates tRNA binding to synthetases. No archaeal orthologs of the two accessory proteins p38 and p43 or the third auxiliary protein p18 have so far been identified. Whether other unidentified proteins exist in Archaea that harbor the same functions as the three mammalian accessory proteins is so far unknown. If the accessory proteins are absent from the archaeal domain, the functions of these factors might instead lie within the synthetases themselves, as suggested for some bacterial aaRS that show similarities in their C- or N-terminal ends with the nonsynthetase factors of the mammalian complex (6). Since LeuRS enhances the catalytic activity of ProRS in M. thermautotrophicus, LeuRS may stabilize the complex, perhaps acting as a scaffold protein. Although no domains similar to the eukaryotic accessory proteins could be detected in archaeal LeuRS or ProRS, both contain unique functional regions that now merit closer investigation with respect to their potential to mediate protein-protein interactions (39–42).
The functional consequences of interactions between aaRSs and other proteins remain unknown in all but a few cases and are generally confined to enhancements in tRNA binding mediated by tRNA-interacting factors (reviewed in Ref. 43). For example, Arc1p/p43 acts in trans to enhance catalytic activity (44, 45), whereas extensions of eukaryotic aaRSs can enhance both specificity (46) and aminoacylation in cis (38). The interaction between LeuRS and ProRS described here falls into neither of these categories, with an aaRS rather than an accessory factor providing catalytic enhancement in trans. Whereas the mechanisms underlying this effect remain unknown, our findings suggest that direct interactions between aaRSs, as seen extensively in higher eukaryotes, could potentially provide a novel means to directly improve aminoacylation efficiency.
Acknowledgments
We thank C. Gruber and M. Smith for making the cDNA library. We thank S. Ataide, J. Levengood, and H. Roy for critical reading of the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM 65183 (to M. I.) and by National Science Foundation Grant MCB-0237483 (to Z. K.).
The abbreviations used are: aaRS, aminoacyl-tRNA synthetase; 3-AT, 3-aminotriazole; HMD, H2-forming N5-N10-methylene tetrahydromethanopterin dehydrogenase; LeuRS, leucyl-tRNA synthetase; ProRS, prolyl-tRNA synthetase; BSA, bovine serum albumin.
M. Prætorius-Ibba and M. Paras, unpublished results.
References
- 1.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Behrens C, Nielsen JN, Fan XJ, Doisy X, Kim KH, Prætorius-Ibba M, Nielsen PE, Ibba M. Tetrahedron. 2000;56:9443–9449. [Google Scholar]
- 3.Mirande, M. (2005) in The Aminoacyl-tRNA Synthetases (Ibba, M., Francklyn, C. S., and Cusack, S., eds) pp. 298–308, Landes Bioscience, Georgetown, TX
- 4.Wolfe CL, Warrington JA, Davis S, Green S, Norcum MT. Protein Sci. 2003;12:2282–2290. doi: 10.1110/ps.03147903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bec G, Kerjan P, Waller JP. J Biol Chem. 1994;269:2086–2092. [PubMed] [Google Scholar]
- 6.Lee SW, Cho BH, Park SG, Kim S. J Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 7.Cerini C, Kerjan P, Astier M, Gratecos D, Mirande M, Semeriva M. EMBO J. 1991;10:4267–4277. doi: 10.1002/j.1460-2075.1991.tb05005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. J Mol Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 9.Quevillon S, Mirande M. FEBS Lett. 1996;395:63–67. doi: 10.1016/0014-5793(96)01005-8. [DOI] [PubMed] [Google Scholar]
- 10.Kim JY, Kang YS, Lee JW, Kim HJ, Ahn YH, Park H, Ko YG, Kim S. Proc Natl Acad Sci U S A. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang J, Kim T, Ko YG, Rho SB, Park SG, Kim MJ, Kwon HJ, Kim S. J Biol Chem. 2000;275:31682–31688. doi: 10.1074/jbc.M909965199. [DOI] [PubMed] [Google Scholar]
- 12.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 13.Deinert K, Fasiolo F, Hurt EC, Simos G. J Biol Chem. 2001;276:6000–6008. doi: 10.1074/jbc.M008682200. [DOI] [PubMed] [Google Scholar]
- 14.Morales AJ, Swairjo MA, Schimmel P. EMBO J. 1999;18:3475–3483. doi: 10.1093/emboj/18.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomanbhoy T, Morales AJ, Abraham AT, Vortler CS, Giege R, Schimmel P. Nat Struct Biol. 2001;8:344–348. doi: 10.1038/86228. [DOI] [PubMed] [Google Scholar]
- 16.Rocak S, Landeka I, Weygand-Durasevic I. FEMS Microbiol Lett. 2002;214:101–106. doi: 10.1111/j.1574-6968.2002.tb11331.x. [DOI] [PubMed] [Google Scholar]
- 17.Dagkessamanskaia A, Martin-Yken H, Basmaji F, Briza P, Francois J. FEMS Microbiol Lett. 2001;200:53–58. doi: 10.1111/j.1574-6968.2001.tb10692.x. [DOI] [PubMed] [Google Scholar]
- 18.Lipman RS, Chen J, Evilia C, Vitseva O, Hou YM. Biochemistry. 2003;42:7487–7496. doi: 10.1021/bi0344533. [DOI] [PubMed] [Google Scholar]
- 19.Goldgur Y, Safro M. Biochem Mol Biol Int. 1994;32:1075–1083. [PubMed] [Google Scholar]
- 20.Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve JN. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiestl RH, Gietz RD. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 22.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 23.Stehlin C, Burke B, Yang F, Liu H, Shiba K, Musier-Forsyth K. Biochemistry. 1998;37:8605–8613. doi: 10.1021/bi980364s. [DOI] [PubMed] [Google Scholar]
- 24.Morgan RM, Pihl TD, Nolling J, Reeve JN. J Bacteriol. 1997;179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman CS, Winston F. Gene (Amst) 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson AJ, Fersht AR, Blow DM, Winter G. Biochemistry. 1983;22:3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- 27.Sampson JR, Uhlenbeck OC. Proc Natl Acad Sci U S A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polycarpo C, Ambrogelly A, Ruan B, Tumbula-Hansen D, Ataide SF, Ishitani R, Yokoyama S, Nureki O, Ibba M, Söll D. Mol Cell. 2003;12:287–294. doi: 10.1016/s1097-2765(03)00280-6. [DOI] [PubMed] [Google Scholar]
- 29.Hou YM, Westhof E, Giege R. Proc Natl Acad Sci U S A. 1993;90:6776–6780. doi: 10.1073/pnas.90.14.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer F, Rohde M, Salzmann M, Jussofie A, Gottschalk G. J Bacteriol. 1988;170:1438–1444. doi: 10.1128/jb.170.4.1438-1444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprott GD, Jarrell KF. Can J Microbiol. 1981;27:444–451. doi: 10.1139/m81-067. [DOI] [PubMed] [Google Scholar]
- 32.Krueger RD, Seeley RJ, Fahrney DE. System Appl Microbiol. 1986;7:388–392. [Google Scholar]
- 33.Reeve JN, Nolling J, Morgan RM, Smith DR. J Bacteriol. 1997;179:5975–5986. doi: 10.1128/jb.179.19.5975-5986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golinelli-Cohen MP, Zakrzewska A, Mirande M. J Mol Biol. 2004;340:15–27. doi: 10.1016/j.jmb.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 35.Norcum MT, Warrington JA. J Biol Chem. 2000;275:17921–17924. doi: 10.1074/jbc.C000266200. [DOI] [PubMed] [Google Scholar]
- 36.Norcum MT, Boisset N. FEBS Lett. 2002;512:298–302. doi: 10.1016/s0014-5793(02)02262-7. [DOI] [PubMed] [Google Scholar]
- 37.Han JM, Kim JY, Kim S. Biochem Biophys Res Commun. 2003;303:985–993. doi: 10.1016/s0006-291x(03)00485-6. [DOI] [PubMed] [Google Scholar]
- 38.Francin M, Kaminska M, Kerjan P, Mirande M. J Biol Chem. 2002;277:1762–1769. doi: 10.1074/jbc.M109759200. [DOI] [PubMed] [Google Scholar]
- 39.Woese CR, Olsen GJ, Ibba M, Söll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukunaga R, Yokoyama S. J Mol Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 41.Ribas De Pouplana L, Brown JR, Schimmel P. J Mol Evol. 2001;53:261–268. doi: 10.1007/s002390010216. [DOI] [PubMed] [Google Scholar]
- 42.Musier-Forsyth, K., Burke, B., and Cusack, S. (2005) in The Aminoacyl-tRNA Synthetases (Ibba, M., Francklyn, C. S., and Cusack, S., eds) pp. 149–161, Landes Bioscience, Georgetown, TX
- 43.Guigou L, Shalak V, Mirande M. Biochemistry. 2004;43:4592–4600. doi: 10.1021/bi036150e. [DOI] [PubMed] [Google Scholar]
- 44.Simos G, Sauer A, Fasiolo F, Hurt EC. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 45.Graindorge JS, Senger B, Tritch D, Simos G, Fasiolo F. Biochemistry. 2005;44:1344–1352. doi: 10.1021/bi049024z. [DOI] [PubMed] [Google Scholar]
- 46.Whelihan EF, Schimmel P. EMBO J. 1997;16:2968–2974. doi: 10.1093/emboj/16.10.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]


